Abstract
English walnut (Juglans regia), has high economic and ecological value. As an important tree species for eliminating poverty, it is planted in many Provinces of China. In 2021, new pathogenic fungi were observed in English walnut in Guangyuan City, Sichuan Province, China. The initial symptom of leaf infection is that the leaves are covered with small black spots, which gradually expand into larger brown spots. Most of the spots appeared at the edges of the leaves, and yellow whorls were observed at the junction between the spots and the healthy leaves. The pathogenic fungi were isoalted form collecting disease samples and purified by single-spore culturing. In vitro and field experiments showed that the pathogen could cause brown spots on walnut leaves. The inoculation experiment showed that the symptoms in the field experiment were the same as those observed on the spot; however, slight differences were observed in the in vitro experiment. Ten isolates were obtained from walnut leaves with brown spot symptoms, and these were further characterized based on morphology and DNA sequencing. ITS (internal transcribed spacer), LSU (large sub-unit rDNA), rpb2 (second largest subunit of RNA polymerase) and tub2 (beta-tubulin) gene regions were used to construct phylogenetic trees and determine the evolutionary relationships among the collected strains. The isolate was identified as Nothophoma quercina by morphological and polygene analyses. As far as we are aware, the brown spots on walnut leaves caused by N. quercina is the first report of its kind.
Similar content being viewed by others
Introduction
English walnut (Juglans regia), belonging to the family Juglandaceae, is a nut tree species whose seeds have high nutritional and economic value and are widely distributed globally1. The J. regia cultivation has a long history in China. This tree species also has a high economic value. The seeds can be used to squeeze extract oil. English walnuts are also becoming increasingly popular due to their high edible value, economic benefits, and adaptability to different soils and climates. With the rapid development of economic forests, walnut planting has become popular in counties, cities, and districts of China. Walnut is the main cash crop in some areas and accounts for approximately 80% of the local income. In 2020, approximately 600, 000 tons of shell walnut seeds were produced in the Sichuan Province. In China, the output and planting area rank third and second, respectively. Owing to the continuous increase in the planting area, the same type of cultivation methods is used under different site conditions and the varieties are frequently introduced from outside the region. Previously rare diseases in this area have become increasingly serious. Different diseases such as anthracnose, branch blight, and walnut rot appear consecutively. In this study, we found that a new disease was emerging. In 2021, walnut leaves with brown spots were found in a walnut garden in Guangyuan City (Sichuan Province, China). Symptoms included a large number of tan spots around the leaves, a black protrusion in the center, and a yellow halo at the junction of the affected part and the healthy leaves. In the late stage, the infected leaves were covered with small pycnidia and turned from dark brown to black, leading to early defoliation. In 2021, nearly 40% of the 50 walnut trees investigated exhibited the same symptoms, causing significant yield loss. Although the yield loss of walnut caused by this pathogen has not been reported yet, the disease causes considerable yield losses in many cherries growing regions2. In recent years, research on walnuts has mainly focused on the optimization of germplasm resources, and approximately 80 cultivars have been generated so far around in Sichuan Province. However, there are few reports on walnut diseases, which are not conducive to the management and operation of the walnut industry in the Sichuan province.
Nothophoma was described by Chen et al.3 There are currently 22 species in this genus. Many species in this genus are plant pathogens, and its members cause diseases in a wide variety of woody plants3,4,5. Several important fruits, including Chaenomeles sinensis, have been reported to be affected by Nothophoma genus fungi in Korea. Nothophoma quercina species are important plant pathogens, have a wide range of economically important plant hosts. The disease symptoms in fruit and nut trees include brown spot of jujube, Aucuba japonica leaf blight, bud blight on Photinia × fraseri, leaf blight of Magnolia coco, leaf spot disease of Phellodendron amurense and trunk canker on Malus micromalus6,7,8,9,10,11. All of these cases were induced by N. quercina. These results confirmed their pathogenicity on hosts as symptoms were reproduced. There have, however, been no reports of walnut brown spots caused by N. quercina. This study aimed to identify the pathogen responsible for walnut brown spot in Sichuan Province, China.
Materials and methods
Sample collection and isolation
The occurrence of walnut brown spot in walnut main producing area of Guangyuan City, Sichuan Province, China was investigated. Guangyuan is located in the northeast of Sichuan Province (36 32′ n, 105 52′E). Located in the south of the Qinling Mountains, this area is the north–south boundary zone of China, and the climate is a typical subtropical humid monsoon climate. The recorded annual average temperature is 16.1 °C and the annual rainfall is 800 to 1000 mm. The main walnut varieties sampled from 6 orchards were “Shuoxing”, “Xiazao”, “Chuanzao 2”, and “Shuchao 2”, with an average orchard age of 14 years. Samples were collected in spring and summer from 2021 to 2022. Fifteen walnut trees of the same age were selected from each orchard. Samples include leaves with obvious disease spots and dead leaves. Plastic Ziploc bags were used to transport samples to the laboratory. A procedure described by Wijesinghe was followed to examine and process the samples12. An NVT-GG dissecting microscope was used to observe and photograph pycnidia external shape, size, and color (Company for cutting-edge photoelectricity technology in Shanghai) VS-800C micro-digital camera matched (Weishen Times Technology Co., Ltd., Shenzhen, China). The microstructure of conidiophores and conidia was observed and photographed using an OLYMPUS BX43 compound microscope and an OLYMPUS DP22 digital camera. Tarosoft® Image Frame Work v.0.9.7 was used to observe and measure the following structures: diameter, height, color, and shape of Conidiophores; length and width of Conidia (Measure the maximum and minimum values and determine the range). Some images are processed by Adobe Photoshop CS6. And describe according to the morphological characteristics. Before separation, wash the blade surface with sterile distilled water for 3 times to remove surface dust. Surface-sterilized in 75% ethanol for 30 s, washed three times with sterile water, and dried at room temperature. Single spore was isolated by microscope and cultured in PDA plate. According to Chomnunti method, pure cultures were obtained from single conidia on potato-dextrose agar (PDA) media13. Cultures were incubated at 25 °C for up to 1 month. Colony characteristics were initially used to identify fungal isolates obtained from walnuts (density, texture, pattern, rate and color of mycelial growth, and presence or absence of pycnidia) and conidial morphology (shape, color, absence or presence of septation of conidia) into Nothophoma genera. The type specimens collected in the field are kept in the Herbarium of Sichuan Agricultural University, Chengdu, China (SICAU). The strains are deposited in the fungal specimen preservation room of Sichuan Agricultural University (SICAUCC).
DNA extraction and polymerase chain reaction (PCR) amplification
DNA was extracted from fresh mycelia with a DNA extraction kit™ (TIANDZ, China), following the specifications of the kit. Operate and use according to the instruction of the reagent. The extracted DNA is stored at 4 °C for normal use, and the long-term storage should be kept at − 20 °C. The primers LR0R and LR7, ITS5 and ITS4, fRPB2-5F and fRPB2-7cR and Bt2a and Bt2b14,15,16,17. Were used for the amplification of the internal transcribed spacers (ITS), large sub-unit rDNA (LSU), the second largest subunit of RNA polymerase II (rpb2) and β-tubulin (tub2), respectively. Polymerase chain reaction (PCR) amplification was carried out following Dai et al.18.
The PCR reaction system was 25 μl, comprises mainly 22 μl of Jingpai PCR mixture (TSINGKE, Chengdu, China), add 1 μl of upstream primer and 1 μl of downstream primer, at last, 1 μl template genomic DNA was added. PCR execution procedure is as follows: LSU, and ITS have the same running procedures. Firstly, denature at 94 °C for 3 min; then denature at 94 °C for 30 s for 35 cycles, annealing at 55 °C for 50 s and elongation at 72 °C for 60 s. Finally, it was extended at 72 °C for 10 min. The following procedures are adopted for rpb2: Firstly, denature at 95 °C for 5 min; then denature at 95 °C for 1 min for 35 cycles, annealing at 52 °C for 2 min and elongation at 72 °C for 90 s; finally, it was extended at 72 °C for 10 min. The following procedures are adopted for tub2: firstly, denature at 94 °C for 3 min; then denature at 94 °C for 3 s for 35 cycles, annealing at 52 °C for 50 s and elongation at 72 °C for 1 min; finally, it was extended at 72 °C for 10 min. The TSINGKE (Chengdu, China) Biological Technology Co., Ltd, was contracted to sequence successfully amplified PCR products. The consensus sequences were obtained from generated sequence reads using Bioedit version 7.0.5.3 software. All the obtained sequences were deposited in NCBI GenBank.
Phylogenetic analysis
The sequences of closely related strains were determined by BLAST search in GenBank. We confirmed and obtained all molecular data of Nothophoma following recent studies8,19. The sequences were downloaded from GenBank. (http://www.ncbi.nlm.nih.gov/). Single gene and phyloygene (LSU, ITS, rpb2 and tub2) Compare all sequences MAFFT v.7 (https://mafft.cbrc.jp/alignment/server/)20. If necessary, use BioEdit v.7.0.5.2 software to make manual improvement21. Manually delete the aligned front and back ends and blurred areas. By comparing the single gene sequence data, the consistency is checked, and the overall topological structure is determined. Polygene sequence consists of Mesquite version 3.11 (build 766)22. Combining the Bayesian inference (BI) and maximum likelihood analysis (ML) analysis, the phylogenetic analysis of ITS, LSU, rpb2 and tub2 gene sequences was carried out. Use Clustal X version 1.81 to convert alignment to NEXUS file (.nxs)23, then BI analysis is carried out. According to the Akaike Information Criterion (AIC), MrModeltest. v.2.2 determined the best nucleotide substitution model24. Maximum likelihood analysis using CIPRES science gateway network server25, and chosen RAxML-HPC2 on XSEDE (8.2.10)26 GTRGAMMA substitution model is used for 1000 boot iterations. Bayesian analysis was performed by MR Bayes v.3.2.227. Run 6 Markov chains at the same time, totaling 5000 generations, and sample the tree every 100 generations. The aging frequency is set to 0.25, and when the average standard deviation of splitting frequency reaches below 0.01, the system will automatically stop running28. Phylogenetic tree was visualized by Fig Tree v.1.4.329. In addition, Adoble Illustrator CS6 v.16.0.0 is used for layout and improvement. The maximum likelihood guidance value (MLBP) is equal to or greater than 63%. Bayesian posterior bootstrap values (BYPP) is greater than 0.54. All sequences used and generated in this paper are submitted to GenBank and listed in Table 1.
Pathogenicity tests
Four isolates were selected for the subsequent pathogenicity test. Before inoculation, the isolates were cultured on Potato Dextrose Agar (PDA) at 25 °C for 15 days under 12 h fluorescent light/dark conditions.
Pathogenicity on walnut plant
The pathogenicity test was performed on six plant 2 year-old potted walnut. The 2-year-old grafted seedlings were collected and cultured in the greenhouse. The spore suspension was then sprayed onto the tender leaves of cuttings from the three seedlings. Sterile water was sprayed on the tender leaves of three grafted seedlings as a control. All six treatments were bagged and the relative humidity was ≥ 90%. The seedlings were kept in a greenhouse for 30 days. Then, the collected healthy young leaves were disinfected in 3% sodium hypochlorite for 60 s and 75% ethanol for 60 s, washed in sterile water 3–5 times, dried in air, and finally inoculated with conidia suspension (4.5 × 105 conidia/mL). During the experiment, 12 healthy young leaves were sprayed with spore suspension, and the remaining 12 leaves were sprayed with sterile water. The inoculated leaves were placed on a flat plate sterilized at high temperature and placed in a constant temperature light incubator at 25 °C (relative humidity 90%) for cultivation and observation. Every 7 day after inoculation, the disease severity was evaluated by the percentage (dpi) by measuring the leaf surface size with disease symptoms. This experiment was repeated three times.
Ethics statement
All plant materials and fungal specimens are treated according to international standards.
Results
Disease symptoms identification
With typical brown necrotic spots (Fig. 1a) a walnut orchard with typical disease spots was observed in the Chaotian District of Guangyuan City (36°32′28″N, 105°52′26″E, 557 m above sea level), Sichuan Province, China. All symptoms were observed in all the six orchards surveyed. These small brown necrotic areas appeared on new leaves, first from the tip and edge of the leaf, and developed inward. The lesion expanded to form a dark brown necrotic area and some leaves had brownish-red edges (Fig. 1a,c). This leads to the distortion and necrosis of the leaves (Fig. 1b). Finally, the leaves died and fell in advance (Fig. 1d,e). In June 2020, a large number of typical symptoms of walnut brown spot were found in the walnut base in Guangyuan City, Sichuan Province. the total planting area of walnut in this base is about 1000 hectares. Ninety samples with typical symptoms were collected from six orchards, and the fungi isolated from the 60 samples were morphologically similar to Nothophoma quercina. According to the statistical analysis of collected samples, the incidence of walnut leaf brown spot is about 40%.
Isolation of fungi
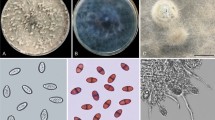
Ten isolates were isolated from diseased leaves with walnut brown spots (Fig. 2a), which were cultured on PDA medium using the same method as Chomnunti et al.13 Conidia germinate in 24 h (Fig. 2g). colonies grow in a circle, with regular edges and wool-like hypha; the aerial hyphae are white at first, aerial hyphae were initially white, after the seventh day, it gradually changed from brown to dark brown at 25 °C under a 12 h fluorescent light/dark regime. The colonies are distributed in a circular cushion shape, and the aerial hyphae are lighter in color (Fig. 2h,i). After 14 ~ 20 days of culture, a large number of dark brown pycnidia were observed on the surface of the culture medium. Pycnidia were measurement (210–)220–360(− 365) × 190(− 195)–(230–)240 μm (\({\overline{\text{x}}}\) ± SD = 290 ± 4 × 205 ± 3 μm, n = 20), drown to dark brown, hemispherical, spherical or irregular. Conidia measurement size (3.7–)4.2–8 × 2.7(– 3)–(4.8–)5.6 μm (\({\overline{\text{x}}}\) ± SD = 6.1 ± 0.6 μm × 4.1 ± 0.4 μm, n = 50), most of them are oval, partially circular, smooth, hyaline, aseptate, thick-walled, Light brown at maturity (Fig. 2b–f). Isolates with small morphological differences were then compared with isolates from different hosts with larger conidia and pycnidia. This morphological similarity was higher than that of the type species. It was found that there was no significant difference in the size of conidia among the isolates. However, the differences of pycnidial size among isolates were obvious. The host plants of Nothophoma quercina from different locations differ, however, no reports have been found on walnut plants.
Phylogenetic analysis
Only morphological identification can no longer meet the needs of current identification. In order to verify the accuracy of our morphological identification, we also added molecular biological identification. So as to determine its pathogen. Gene fragments of LSU, ITS, tub2 and rpb2 in Nothophoma quercina (SICAUCC 22-0080, SICAUCC 22-0081, SICAUCC 22-0082 and SICAUCC 22-0083) were then amplified and sequenced. Search results showed that the homology of each gene sequence (LSU, ITS, tub2 and rpb2) of 10 isolates was 100%. And the target sequences of LSU, ITS, tub2 and rpb2 of four representative isolates were selected. (SICAUCC 22-0080, SICAUCC 22-0081, SICAUCC 22-0082, SICAUCC 22-0083) all sequences are stored in GenBank (Login number: ON707468, ON707463, ON645227, ON645232; ON707469, ON707464, ON645228, ON645233; ON707470, ON707465, ON645229, ON645234; ON707471, ON707466, ON645230, ON645235, respectively). All the population sequences and close species of this genus were selected from this genus, and a total of 73 species were used to construct phylogenetic tree. The sequence contains a combined data set of LSU, ITS, tub2 and rpb2 sequences, which contains 3882 base pairs (bp). Firstly, the maximum-likelihood method is used to analyze phylogeny. Among them, Leptosphaeria doliolum and L.conoidea were used as external groups for phylogenetic analysis. Phylogenetic tree unites Nothophoma genera on the same major clade, four new sequences, SICAUCC 22–0080, SICAUCC 22-0081, SICAUCC 22-0082, SICAUCC 22-0083, are gathered in a well-supported Nothophoma quercina (ML = 98%). The results of ML phylogenetic analysis are shown in Fig. 3. Then, the phylogenetic tree is constructed by BI, and the tree shape is the same as ML tree. Isolates SICAUCC 22-0080, SICAUCC 22-0081, SICAUCC 22-0082 and SICAUCC 22–0083 were clustered in this N. quercina, which supported the value (ML/BI = 98/1), including N. quercina (CBS 633.92) in type species. Four isolates were simultaneously identified as Nothophoma quercina (Syd.) Q. Chen & L. Cai. This is the result of the joint identification of phylogenetic trees based on morphological characteristics, cultural characteristics and LSU ITS, rpb2 and tub2 sequence analysis.
RAxML tree based on a combined dataset of a partial LSU, ITS, tub2 and rpb2 sequence analysis in Didymellaceae. The tree is rooted with Leptosphaeria conoidea (CBS 616.75) and L. doliolum (CBS 505.75). ML and Bayesian posterior probabilities values are given at the nodes. The new isolate is highlighted in bold.
Pathogenicity tests
The pathogenicity of the tender leaves of living plants was verified in vitro. On the tender leaves that had been detached, conidial suspension (4.5 × 105 conidia/mL) was sprayed onto them. The results showed that the tender leaves in vitro appeared as typical leaf necrosis with light brown to brown spots edge 15 days after inoculation (Fig. 4b1–b3), whereas the control plants were still asymptomatic (Fig. 4a1–a3). After inoculation of healthy walnut seedling leaves for 20 days, the symptoms were the same as those observed in the field (Fig. 5a1–a3), whereas the control plants were still asymptomatic (Fig. 5b1–b3). Fungi were re-isolated from all infected leaves, which were similar to Nothophoma quercina by comparison of culture characteristics and morphology. The re-isolated strain was identified as Nothophoma quercina by sequencing analysis following Koch’s postulate.
Discussion
Nothophoma quercina is the first confirmed species in this genus, thus confirming the status of Nothophoma. N. quercina was discovered and named by Chen et al.2. This species was originally identified as Cicinobolus quercinus, and then transferred to Ampelomyces genus. Later, Aveskamp et al.30 regarded it as one of Phoma and renamed it. Phylogenetic analysis of multiple genes showed that this species gathered in in the Nothophoma. Finally, it is proposed that N. quercina as a new combination creates Nothophoma genus. To date, 22 species have been identified, including N. anigozanthi, N. arachidis-hypogaeae, N brennandiae, N. chromolaenae, N. eucalyptigena, N. acaciae, N. ferruginea, N. macrospora, N. pruni, N. infossa, N. infuscata, N. multilocularis, N. garlbiwalawarda, N. naiawu, N. nullicana, N. prosopidis, N. pruni, N. spiraeae, N. quercina, N. raii, and N. variabilis, N. gossypiicola3,4,18,31,32,33,34,35,36,37.
In this study, molecular verification of Nothophoma quercina was performed using polygene phylogenetic analysis. For the identification of the pathogen of walnut brown spot caused by N. quercina in Sichuan, China, this experiment adopted the combination of morphology and molecule for the first time. The morphological characteristics of the isolated strains are consistent with those described by N. quercina. However, due to different nutritional conditions and temperature changes, there are slight differences among different strains. Identification of pathogen of walnut leaf spot is the key of successful control of this disease. Therefore, it is extremely important to correctly identify pathogenic fungi.
Nothophoma quercina shoot blight, causes cankers. Among them, there are many reports on leaf diseases of various plants, including Garryaceae, Rhamnaceae, Rosaceae, Rhamnaceae, Anacardiaceae, Oleaceae, Ulmaceae and Fagaceae. N. quercina is morphologically and geographically diverse. This species has been found in Aucuba japonica (China), Ziziphus jujuba (China), Magnolia coco (China), Pseudocydonia sinensis (Korea), Chaenomeles sinensis (Korea), and Prunus dulcis (Tunisia)6,7,8,38,39,40. In the world, we first discovered the brown spot of walnut leaves caused by N. quercina.
Walnut is an important economic tree species, oil tree species, and ecological tree species worldwide41. Walnut is widely planted all over the world, especially in Europe, Asia and many parts of America42. Because it is loved by people, it is called one of the four dried fruits, along with cashew nut, hazelnut and almond. The walnut germplasm resources are abundant in China. At present, 13 species of Juglans have been identified, with more than 300 cultivated varieties. Among them, Juglans regia and J. sigillata have the largest planting areas43. However, the aggravation of diseases and insect pests is constantly affecting the healthy development of walnut industry. In particular, Nothophoma quercina induced leaf diseases lead to early defoliation and fruit loss of J. regia. N. quercina is a common pathogen in plants. Plant infections are gradually increasing and the scope of damage is constantly expanding. Therefore, it is important to investigate this disease. In this study, the pathogen was identified by morphological and molecular biology methods. Finally, it was verified by pathogenicity test. But these are only the first steps of disease control. At present, further research is being carried out to obtain the mode of occurrence and development of diseases. Understand when and how pathogens infect plants in the growing season, and find out all kinds of environmental factors conducive to disease development. Precise information regarding the disease cycle and epidemiology is required to provide walnut growers with effective management recommendations. Get accurate information about the epidemic of diseases, and provide effective management suggestions for walnut growers according to the information.
Data availability
The data presented in this study are deposited in the NCBI GenBank (accession numbers ON707468, ON645232, ON707463, ON645227; ON707469, ON645233, ON707464, ON645228; ON707470, ON645234, ON707465, ON645229; and ON707471, ON645235, ON707466, ON645230.
References
Liu, M. et al. Walnut fruit processing equipment: Academic insights and perspectives. Food Eng. Rev. 13(4), 822–857. https://doi.org/10.1007/s12393-020-09273-6 (2021).
Chethana, K. et al. Molecular characterization and pathogenicity of fungal taxa associated with cherry leaf spot disease. Mycosphere 10(1), 490–530. https://doi.org/10.5943/mycosphere/10/1/8 (2019).
Chen, Q., Jiang, J. R., Zhang, G. Z., Cai, L. & Crous, P. W. Resolving the Phoma enigma. Stud. Mycol. 82, 137–217. https://doi.org/10.1016/j.simyco.2015.10.003 (2015).
Zhang, L. X., Yin, T., Pan, M., Tian, C. M. & Fan, X. L. Occurrence and identification of Nothophoma spiraeae sp. nov. in China. Phytotaxa 430(3), 147–156. https://doi.org/10.11646/phytotaxa.430.3.1 (2020).
Gomzhina, M., Gasich, E., Khlopunova, L. & Gannibal, P. New species and new findings of Phoma-like fungi (Didymellaceae) associated with some Asteraceae in Russia. Nova Hedwigia 111(1–2), 131–149. https://doi.org/10.1127/novahedwigia/2020/0586 (2020).
Bai, J. Y., Wang, X. M., Shi, Y. J. & Duan, C. X. Occurrence and identification of Nothophoma quercina causing brown spot of jujube in China. Can. J. Plant Pathol. 38, 527–532. https://doi.org/10.1080/07060661.2016.1266389 (2016).
Lv, Y. C. et al. First report of leaf blight on Aucuba japonica caused by Nothophoma quercina in China. Plant Dis. 104(10), 2731–2731. https://doi.org/10.1094/PDIS-01-20-0201-PDN (2020).
Zou, J., Dong, Y., Wang, H., Liang, W. & Li, D. Identification and Characterization of Nothophoma quercina causing bud blight on Photinia × fraseri in China. Plant Dis. 105(5), 1356–1364. https://doi.org/10.1094/PDIS-06-20-1218-RE (2021).
Zeng, Q. et al. First report of leaf blight on Magnolia coco caused by Nothophoma quercina in China. Plant Dis. 106, 761–761. https://doi.org/10.1094/PDIS-05-21-1061-PDN (2021).
Jiao, W. L., Zhou, R. J., Fu, J. F., Xu, H. J. & Hao, N. First report of Nothophoma quercina causing leaf spot disease of Phellodendron amurense in China. Plant Dis. 101(10), 1820. https://doi.org/10.1094/PDIS-01-17-0079-PDN (2017).
Liu, M. et al. First report of Nothophoma quercina causing trunk canker on crabapple (Malus micromalus) in China. Plant Dis. 102(7), 1462–1462. https://doi.org/10.1094/PDIS-11-17-1866-PDN (2018).
Wijesinghe, S. N., Dayarathne, M. C., Maharachchikumbura, S. S., Wanasinghe, D. N. & Hyde, K. D. Ceramothyrium chiangraiense, a novel species of Chaetothyriales (Eurotiomycetes) from Ficus sp. in Thailand. Asian J. Mycol. 2, 269–280. https://doi.org/10.5943/ajom/2/1/17 (2019).
Chomnunti, P. et al. The sooty moulds. Fungal Divers. 66(1), 1–36. https://doi.org/10.1007/s13225-014-0278-5 (2014).
Liu, Y. J., Whelen, S. & Hall, B. D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092 (1999).
Vilgalys, R. & Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990 (1990).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (eds Innis, M. A. et al.) 315–322 (Academic Press, 1990). https://doi.org/10.1016/B978-0-12-372180-8.50042-1.
Glass, N. L. & Donaldson, G. C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. https://doi.org/10.1128/AEM.61.4.1323-1330.1995 (1995).
Dai, D. Q. et al. Bambusicolous fungi. Fungal Divers. 82, 1–105. https://doi.org/10.1007/s13225-016-0367-8 (2017).
Keirnan, E. C. et al. Cryptic diversity found in Didymellaceae from Australian native legumes. MycoKeys 78, 1–20. https://doi.org/10.3897/mycokeys.78.60063 (2021).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30(4), 772–780. https://doi.org/10.1093/molbev/mst010 (2013).
Hall, T. BioEdit. Ibis Therapeutics, Carlsbad, CA, 92008, USA. http://www.mbio.ncsu.edu/BioEdit/bioedit.html (2004).
Maddison, W.P. and Maddison, D.R. Mesquite: A modular system for evolutionary analysis, Version 3.61. http://www.mesquiteproject.org (2019).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. https://doi.org/10.1093/nar/25.24.4876 (1997).
Nylander, J.A.A. MrModeltest 2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University (2004).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE) (eds Miller, M. A. et al.) 1–8 (IEEE, 2010). https://doi.org/10.1109/GCE.20.
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. https://doi.org/10.1093/bioinformatics/btu033 (2014).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. https://doi.org/10.1093/bioinformatics/btg180 (2003).
Maharachchikumbura, S. S., Hyde, K. D., Gareth Jones, E. B., Mckenzie, E. H. C. & Huang, S.-K. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 72, 199–301. https://doi.org/10.1007/s13225-015-0331-z (2015).
Rambaut, A. and Drummond A. FigTree: Tree figure drawing tool, version 1.4.3. http://tree.bio.ed.ac.uk/software/figtree/ (2016).
Aveskamp, M. M., De Gruyter, J., Woudenberg, J. H., Verkley, G. J. & Crous, P. W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol. 65, 1–60. https://doi.org/10.3114/sim.2010.65.01 (2010).
Yun, Y. H. & Oh, M. H. First report of Nothophoma quercina causing shoot canker on Chaenomeles sinensis in Korea. Plant Dis. 100(12), 2533. https://doi.org/10.1094/PDIS-06-16-0870-PDN (2016).
Hou, L. W. et al. The phoma-like dilemma. Stud. Mycol. 96, 309–396. https://doi.org/10.1016/j.simyco.2020.05.001 (2020).
Mapook, A. et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 101(1), 1–175. https://doi.org/10.1007/s13225-020-00444-8 (2020).
Zhang, Y. et al. Toward a natural classification of Botryosphaeriaceae: A study of the type specimens of Botryosphaeria sensu lato. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.737541 (2021).
Abdel-Wahab, M. A., Bahkali, A. H., El-Gorban, A. M. & Hodhod, M. S. Natural products of Nothophoma multilocularis sp. nov. an endophyte of the medicinal plant Rhazya stricta. Mycosphere 8, 1185–1200. https://doi.org/10.5943/mycosphere/8/8/15 (2017).
Chethana, K. W. T. et al. Molecular characterization and pathogenicity of fungal taxa associated with cherry leaf spot disease. Mycosphere 10(1), 490–530. https://doi.org/10.5943/mycosphere/10/1/8 (2019).
Crous, P. W. et al. Fungal planet description sheets: 625–715. Persoonia 39, 270–467. https://doi.org/10.3767/persoonia.2017.39.11 (2017).
Bensch, K. et al. Cladosporium species in indoor environments. Stud. Mycol. 89, 177–301. https://doi.org/10.1016/j.simyco.2018.03.002 (2018).
Kazerooni, E. A., Maharachchikumbura, S. S. N. & Kang, S. M. First report of fruit canker caused by Nothophoma quercina on Chinese quince in South Korea. Plant Dis. 105(11), 3760–3760. https://doi.org/10.1094/PDIS-12-20-2583-PDN (2021).
Triki, M. A. et al. First report of branch blight of almond trees caused by Nothophoma quercina in Tunisia. J. Plant Pathol. 101(4), 1277–1277. https://doi.org/10.1007/s42161-019-00342-2 (2019).
Wang, Q. H. et al. Walnut anthracnose caused by Colletotrichum siamense in China. Australas. Plant Pathol. 46, 585–595. https://doi.org/10.1007/s13313-017-0525-9 (2017).
Bernard, A., Lheureux, F. & Dirlewanger, E. Walnut: Past and future of genetic improvement. Tree Genet. Genomes https://doi.org/10.1007/s11295-017-1214-0 (2018).
Yuan, X. Y. et al. Population structure, genetic diversity, and gene introgression of two closely related walnuts (Juglans regia and J. sigillata) in Southwestern China revealed by EST-SSR markers. Forests https://doi.org/10.3390/f9100646 (2018).
Acknowledgements
This study was supported by the Sichuan Science and Technology Program under Grant 2022NSFSC1011.
Author information
Authors and Affiliations
Contributions
Y.G.L. and C.L.Y. designed the investigation. C.L.Y. contributed for the research fund. F.H.W., C.L, Q.Z., Y.J.Z. and F.L. performed the sample collection and handling and the literature study. F.H.W., X.L.X. and H.B.Y. conducted the preliminary analysis and pathogenicity test. F.H.W. analyzed the data and wrote the manuscript. C.L.Y. revised and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, F., Liu, C., Zeng, Q. et al. Identification and pathogenicity analysis of leaf brown spot of Juglans regia in China. Sci Rep 13, 6599 (2023). https://doi.org/10.1038/s41598-023-33853-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-33853-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.