Abstract
Evolution of pest resistance reduces the benefits of widely cultivated genetically engineered crops that produce insecticidal proteins derived from Bacillus thuringiensis (Bt). Better understanding of the genetic basis of pest resistance to Bt crops is needed to monitor, manage, and counter resistance. Previous work shows that in several lepidopterans, resistance to Bt toxin Cry2Ab is associated with mutations in the gene encoding the ATP-binding cassette protein ABCA2. The results here show that mutations introduced by CRISPR/Cas9 gene editing in the Helicoverpa zea (corn earworm or bollworm) gene encoding ABCA2 (HzABCA2) can cause resistance to Cry2Ab. Disruptive mutations in HzABCA2 facilitated the creation of two Cry2Ab-resistant strains. A multiple concentration bioassay with one of these strains revealed it had > 200-fold resistance to Cry2Ab relative to its parental susceptible strain. All Cry2Ab-resistant individuals tested had disruptive mutations in HzABCA2. We identified five disruptive mutations in HzABCA2 gDNA. The most common mutation was a 4-bp deletion in the expected Cas9 guide RNA target site. The results here indicate that HzABCA2 is a leading candidate for monitoring Cry2Ab resistance in field populations of H. zea.
Similar content being viewed by others
Introduction
Genetically engineered crops that produce insecticidal proteins from Bacillus thuringiensis (Bt) are useful for managing some economically important pests. Benefits of Bt crops include better pest control and decreased use of conventional insecticides, which enhances yields and farmer profits, and confers health and environmental benefits1,2,3,4,5,6. However, the evolution of resistance to Bt crops by pests decreases these benefits7. Understanding the genetic basis of resistance to Bt crops can improve the monitoring and management of resistance, as well strategies to counter pest resistance.
Some ATP-binding cassette (ABC) transporter proteins mediate Bt intoxication in insects8,9,10. Natural mutations in ABCC2 are genetically linked with resistance to several Bt toxins in the Cry1 family (Cry1Ab, Cry1Ac, and/or Cry1Fa) in seven lepidopterans11,12,13,14,15,16. Likewise, resistance to Cry2Ab is linked with natural mutations in another ABC transporter gene, ABCA2, in Helicoverpa armigera17, Helicoverpa punctigera17, Trichoplusia ni18, and in lab- and field-selected populations of Pectinophora gossypiella19,20. Furthermore, knockout mutations in ABCA2 generated in the laboratory using CRISPR/Cas9 or TALENs caused resistance to Cry2Ab in H. armigera21, T. ni18, Bombyx mori22, and P. gossypiella23. These results provide compelling evidence that ABCA2 plays an important role in the mode of action of Cry2Ab in these lepidopterans.
Here we tested the hypothesis that CRISPR/Cas9-mediated disruption of the gene encoding ABCA2 can cause resistance to Cry2Ab in Helicoverpa zea (also known as corn earworm and bollworm). This lepidopteran is a major pest of many crops in the New World, including corn, cotton, soybean, and sorghum4,24. It has evolved practical resistance to Bt corn and cotton producing Cry1 and Cry2A toxins throughout much of the southern United States7,25,26,27,28,29,30,31,32. Also, early signs of H. zea evolution of resistance to Vip3Aa produced by Bt crops have been reported29,33. Hence, strategies to delay or overcome the evolution of Bt resistance in H. zea are urgently needed. Although timely identification of genes causing Bt resistance remains challenging, new tools have spurred progress with H. zea and H. armigera34,35,36.
In this study, we discovered that introducing disruptive CRISPR/Cas9-mediated mutations in the HzABCA2 gene in a susceptible strain caused resistance to Cry2Ab. We also used CRISPR/Cas9 to introduce disruptive mutations in the HzTO gene (also known as tryptophan 2,3-oxygenase or vermilion), which confirmed previous results showing this causes yellow eye color in H. zea37. All Cry2Ab-resistant individuals tested were found to harbor disruptive mutations in HzABCA2 DNA in the single-guide RNA (sgRNA) target sites used for gene editing. These findings indicate that ABCA2 is important in toxicity of Cry2Ab to H. zea.
Results
In vitro Cas9 cleavage of HzABCA2 PCR products with guide RNAs
We tested seven HzABCA2 sgRNAs separately by combining the sgRNA/Cas9 ribonucleotide mixture for each sgRNA (#1–7) with gDNA amplified by PCR from HzABCA2. Cas9 cleavage of the HzABCA2 amplicons in vitro occurred with sgRNA 7, but not sgRNAs 1–6 (Supplementary Fig. S1).
Creation of Cry2Ab-resistant strains Hz-R2 and Yellow-R2
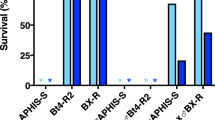
For our experimental design, we targeted HzABCA2 to assess its role in Cry2Ab resistance and HzTO as a secondary and visible marker for gene editing. Of 177 embryos (G0) from the susceptible LAB-S strain injected with the ribonucleotide mixture containing HzTO sgRNA and HzABCA2 sgRNA 7, 97% hatched and 73% pupated. To start the Hz-R2 strain, we pooled 55 G0 pupae (one cup with 10 males + 10 females, one cup with 10 males + 9 females, and one cup with 8 males + 8 females) and allowed the adults to eclose and mate. We established the Yellow-R2 strain by pairing two G0 adult females that had yellow eyes (Supplementary Fig. S2) with two Hz-R2 G0 males having mosaic green/yellow eyes and allowing them to propagate. We screened G1 neonates from both Hz-R2 and Yellow-R2 on diet containing the diagnostic concentration of 1 μg Cry2Ab per cm2 diet. Survival was 58% for Yellow-R2 (n = 224) and 93% for Hz-R2 (n = 192), suggesting that CRISPR/Cas9 efficiently introduced germline mutations in HzABCA2 that caused resistance to Cry2Ab. Survival was 0% for LAB-S (n = 32), which is significantly lower than survival of the G1 larvae from either Hz-R2 or Yellow-R2 (Fisher’s exact test, P < 0.0001 for each comparison). In contrast, survival on control diet did not differ significantly between LAB-S (91%) and either Hz-R2 (88%) or Yellow-R2 (100%) (Fisher’s exact test, P > 0.5 in each comparison). We used the G1 survivors of exposure to Cry2Ab to continue the Yellow-R2 and Hz-R2 strains. In effect, this exposure constituted a single selection for resistance to Cry2Ab after which both strains were reared without exposure to the toxin.
Analysis of the G4 larvae from Hz-R2 revealed they were highly resistant to Cry2Ab relative to their parent strain LAB-S (Fig. 1). Survival for Hz-R2 was 96% for larvae exposed to diet with 30 μg Cry2Ab per cm2 diet (the highest concentration tested). The concentration of Cry2Ab killing 50% of larvae (LC50 in μg Cry2Ab per cm2 diet) was > 30 for the G4 larvae from Hz-R2 and 0.14 (95% CI 0.10–0.19) for LAB-S, which yields a resistance ratio of > 217 for Hz-R2 relative to LAB-S.
To initiate Yellow-R2 containing the desired eye phenotype, we sorted G2 pupae by sex, selected adults with yellow eyes (14 males + 14 females) and allowed them to mate. After six generations of laboratory rearing without additional selection for yellow eyes, all 1,200 individuals examined from the G7-9 Yellow-R2 strain had yellow eyes (n = 400 per generation). This suggests that a HzTO mutation responsible for yellow eyes was likely fixed in the Yellow-R2 strain.
Mutations in sgRNA target sites in HzABCA2 and HzTO gDNA from the Yellow-R2 and Hz-R2 strains
For HzABCA2, we sequenced two clones corresponding to exon 4 (containing HzABCA2 sgRNA 7) from three Hz-R2 and Yellow-R2 G1 survivors reared on diet treated with 1 μg Cry2Ab per cm2 (total of 12 clones) and three LAB-S individuals on untreated diet (total of six clones; Fig. 2). All 12 clones from Hz-R2 and Yellow-R2 had mutations in the HzABCA2 sgRNA 7 target site (Fig. 2A) which introduce premature stop codons (Fig. 2B; Supplementary Fig. S3). In total, we found five unique mutations in HzABCA2 from Hz-R2 and Yellow-R2: a 1-bp deletion at base 867, a 4-bp deletion at position 875–878, an A869C substitution, an A874T substitution, and a 1-bp insertion after base 874. The most common mutation was the 4-bp deletion (bases 875-CAGT-878), which occurred in five of six Hz-R2 clones and all six Yellow-R2 clones. No mutations were found in exon 4 of HzABCA2 in the six clones sequenced from LAB-S.
Mutations in HzABCA2 gDNA and the resulting translated sequences from three Hz-R2 larvae and three Yellow-R2 that survived exposure to diet treated with 1 μg Cry2Ab per cm2 diet. (A) Partial HzABCA2 genomic DNA fragments corresponding to HzABCA2 sgRNA 7 were cloned and Sanger sequenced. Two clones from each of three Hz-R2 larvae and three Yelllow-R2 larvae (2 clones × 3 individuals) and two clones from three LAB-S moths were sequenced. HzABCA2 sgRNA 7 sequence is bold and underlined. The PAM sequence is highlighted in pink. The DNA insertion is shown with red text, deletions are highlighted in blue, and substitutions are highlighted in yellow. Numbers indicate nucleotide position in the HzABCA2 mRNA because only the exon section is shown. (B) Alignment of translated HzABCA2 partial amino acid sequences from the Yellow-R2 and Hz-R2 G1 survivors on 1 μg Cry2Ab per cm2. Amino acids highlighted in green correspond to the gDNA Sanger sequenced for the corresponding HzABCA2 sgRNA 7 target site. Stars indicate premature stop codons. Numbers indicate the position of the amino acid in the HzABCA2 protein sequence.
For HzTO, we sequenced two clones corresponding to exon 6 (the site of the HzTO sgRNA) from four Yellow-R2 G1 moths that had yellow eyes (total of 8 clones) and two clones from one LAB-S moth. Whereas both clones from LAB-S had the wild-type HzTO sequence, all eight clones from Yellow-R2 harbored disruptive mutations (Supplementary Fig. S4A). Overall, we found three different mutations in the HzTO gDNA from Yellow-R2, including a 2-bp insertion in one clone, a 7-bp deletion in five clones, and a 33-bp insertion in two clones (Supplementary Fig. S4A). All three CRISPR/Cas9-induced mutations disrupt the HzTO coding sequence with premature stop codons. The 7-bp deletion and the 2-bp insertion introduced a premature stop at the same codon position in HzTO (corresponding to bases 831 and 840, respectively) and the 33-bp insertion introduced a stop codon in the final 3 bp of the inserted sequence (Supplementary Figs. S4B, S5).
Inheritance of resistance to Cry2Ab in strain Hz-R2
To evaluate inheritance of resistance to Cry2Ab, we used diet overlay bioassays to test Hz-R2, LAB-S, and their F1 progeny resulting from mass crosses. Responses of the F1 progeny show that resistance to Cry2Ab in Hz-R2 was autosomal and recessive (Fig. 3). At the diagnostic concentration, survival was 0% for LAB-S, 100% for Hz-R2, and 0.4% for the F1 progeny (pooled from the two Hz-R2 X LAB-S reciprocal crosses).
The dominance parameter (h), which varies from 0 for completely recessive resistance to 1 for completely dominant resistance, was determined for the F1 progeny from pooled Hz-R2 X LAB-S mass crosses. The results yield h = 0.0041, indicating recessive inheritance of resistance for Hz-R2 at 1 μg Cry2Ab per cm2 diet.
Discussion
The results here show that introduction of disruptive mutations in the H. zea ABCA2 gene (HzABCA2) via CRISPR/Cas9 gene editing caused resistance to Cry2Ab. This finding is consistent with previous results from editing the homologous gene in H. armigera, T. ni, B. mori, and P. gossypiella18,21,22,23. Collectively these data indicate that ABCA2 is important for toxicity of Cry2Ab in at least five species representing three families of Lepidoptera (Noctuidae, Bombycidae, and Gelechiidae). Despite its evident functional importance, the precise role of this protein in the mode of action of Cry2Ab remains obscure10. Heckel (2021) has proposed that elucidating the three-dimensional structures of the ABC transporters will be critical for progress in understanding how these proteins facilitate pore formation by Bt toxins.
Here, we determined that the Hz-R2 strain of H. zea, which harbored CRISPR/Cas9-induced mutations in HzABCA2, was > 200-fold resistant to Cry2Ab compared to its susceptible parental strain LAB-S. Similarly, CRISPR/Cas9 knockout of ABCA2 in two H. armigera lab strains from China revealed Cry2Ab resistance ratios of > 100 relative to the susceptible parental SCD strain21.
The Cry2Ab resistance in lab-selected strains of H. armigera and H. punctigera from Australia is linked with naturally occurring mutations in ABCA217. One of the Cry2Ab-resistant strains of H. armigera from Australia called SP15 was generated from an F2 screen conducted in 200238. Larval survival (adjusted for control mortality) at the highest concentration of Cry2Ab tested was 96% for Hz-R2 and 96% for SP1538. The highest concentration tested was 1.6 times higher for Hz-R2 than SP15 (30 versus 19 μg Cry2Ab per cm2 diet), which means the concentration killing 4% of larvae was higher for Hz-R2 than SP15. In the Australian study38, the mean LC50 (in μg Cry2Ab per cm2 diet) of two unrelated susceptible strains of H. armigera was 0.13, which is nearly identical to the LC50 of 0.14 for the susceptible LAB-S strain tested in this study. Thus, the resistance to Cry2Ab relative to conspecific susceptible strains was similar or slightly greater for the CRISPR-generated mutations in Hz-R2 than for the naturally occurring mutations in SP15. Consistent with the results for Hz-R2, resistance to Cry2Ab was recessive in strains of H. armigera from China and Australia as well as in H. punctigera17,21,38.
Whereas our study using CRISPR/Cas9 and related gene editing studies can rigorously test if mutations in specific genes confer resistance, they cannot determine if mutations in the genes evaluated actually occur in field-selected resistant populations of pests. Accordingly, a key question related to this study is the extent to which mutations disrupting ABCA2 contribute to field-evolved resistance to Cry2Ab. In P. gossypiella from India, mutations disrupting ABCA2 were associated with field-selected resistance to Cry2Ab19,20. In H. armigera and H. punctigera from Australia, mutations disrupting ABCA2 were associated with resistance to Cry2Ab in lab-selected strains derived from field populations17. However, these naturally occurring mutations affecting ABCA2 were not common in field populations of either species in Australia17, which remain susceptible to Cry2Ab39.
The results here showing that mutations disrupting ABCA2 can cause resistance to Cry2Ab in H. zea make it especially compelling to determine if such mutations are associated with the extensive field-evolved resistance of this pest to Cry2Ab in the southern United States7,27,28,29,30,31,32. In a single-pair family established from H. zea collected from corn producing Cry1A.105 + Cry2Ab in Maryland in 2019, resistance to these two Bt toxins was not associated with chromosome 17 where HzABCA2 occurs40. More extensive screening of H. zea from field populations resistant to Cry2Ab is needed to determine if mutations disrupting ABCA2 are important in this widespread practical resistance.
Methods
Insects
We bought H. zea eggs from Benzon Research (Carlisle, PA) in July 2021. We maintained this laboratory strain without exposure to Bt toxins and refer to it as LAB-S because of its susceptibility to Cry2Ab and other Bt toxins27,28,31,41. Larvae were reared on Southland diet (Southland Products, Inc., Lake Village, AR). All rearing and diet bioassays were done at 28 °C under 20–40% humidity and 14 h light:10 h dark. Moths were reared in separate incubators from larvae and had access to a 10% sugar water solution for feeding and cheese cloth for oviposition41.
Bt toxin
We used Cry2A.127, a variant of Cry2Ab protoxin that was prepared, purified, and solubilized as described previously42 and provided by Corteva Agriscience. Cry2A.127 is 98.6% identical with Cry2Ab1 and Cry2Ab2 (9 substitutions out of 633 amino acids) and is referred to here as Cry2Ab.
Design and synthesis of single guide RNA (sgRNA)
We designed sgRNAs targeting two genes: (1) HzABCA2 encoding the ATP-binding cassette protein ABCA2 (from the H. zea genome36); and (2) vermilion (also known as tryptophan 2,3-oxygenase or HzTO) MG976796.1. The latter gene was previously shown to result in yellow mutant eye color in CRISPR/Cas9 mutant knockouts of H. zea37. One sgRNA located in HzTO exon 6 and seven sgRNAs located within 5' exons of HzABCA2 were made (HzABCA2 sgRNAs 1 and 2 in exon 1; sgRNAs 3 and 4 in exon 2; sgRNA 5 in exon 3; and sgRNAs 6 and 7 in exon 4) (Supplementary Table S1). Each sgRNA was adjacent to a corresponding “NGG” PAM site and had no off-target sites based on CRISPOR alignment with H. armigera ABCA2 and TO. Each selected sgRNA target sequence was further checked for potential off-target sites by BLAST searching of the GenBank non-redundant database. sgRNA DNA templates were synthesized as gBlock DNA (Integrated DNA Technologies, Coralville, Iowa) that contained the T7 RNA polymerase binding site, a 20-bp gene-specific target sequence, and the 80-bp common stem-loop tracrRNA sequence. DNA templates were used to synthesize sgRNA using the HiScribe T7 High Yield RNA synthesis Kit (New England Biolabs, Ipswich, MA). Transcribed sgRNAs were treated with DNase I for 20 min at 37 °C and purified using RNAClean XP (Thermo Fisher Scientific) following the manufacturer’s protocol.
In vitro Cas9 cleavage with guide RNAs
To test if each sgRNA complexed with Cas9 was able to cut PCR-amplified HzABCA2 gDNA in vitro, we used the Guide-it sgRNA Screening Kit (Takara Bio, Mountain View, CA) as previously published23. We first extracted gDNA from LAB-S 3rd instar larvae using the Gentra Puregene Tissue Kit (Qiagen, Hilden, Germany), which served as DNA template for PCR. Primer pairs 1HzABCA2-5 + 2HzABCA2-3, 3HzABCA2-5 + 4HzABCA2-3, and 5HzABCA2-5 + 6HzABCA2-3 (Supplementary Table S2) were used to amplify PCR products corresponding to exon 1, exon 2, and exons 3–4, respectively. Each sgRNA was diluted to 50 ng μL−1 in RNase-free water and 1 μL was mixed with 250 ng of Guide-it Cas9 nuclease and incubated at 37 °C for 5 min. Fifty ng of each gDNA PCR template was combined with Cas9 Reaction Buffer, bovine serum albumin, RNase-free water, and the appropriate Cas9/sgRNA mixture. These were incubated at 37 °C for 1 h. Reactions were terminated (80 °C for 5 min) and aliquots of each digestion reaction and corresponding negative controls were analyzed by 1.5% agarose gel electrophoresis stained with SYBR Safe DNA Gel Stain (Thermo Fisher Scientific).
Embryo microinjection
sgRNA targeting HzABCA2 and HzTO were singly complexed with the Alt-R Streptococcus pyogenes HiFi Cas9 nuclease V3 (Integrated DNA Technologies), at 50 ng µL−1 of sgRNA to 100 ng µL−1 of Cas9, then incubated at room temperature for 15 min to generate the Cas9-ribonucleoprotein (RNP) complex. The two RNP complexes were combined to create a mixture of HzABCA2 and HzTO RNP, then the mixture was placed on ice and immediately used for injections.
Embryos were collected from LAB-S by placing microscope glass coverslips (24 X 40 mm, Corning Inc., Corning, NY) on top of screened lids for several cages containing 5 male and 5 female adults for 45 min. Adult H. zea females naturally affix eggs to the coverslips and no additional manipulation of the embryos was needed. An IM-300 microinjector (Narishige International USA, Amityville, NY) equipped with an Olympus IMT-2 inverted microscope (Olympus Corporation, Center Valley, PA) was used to inject newly laid eggs (less than 1 h old) with approximately 100–200 picoliters of the HzTO + HzABCA2 RNP solution. Quartz needles (Sutter Instrument Co, Novato, CA) were beveled using a Model EG-44 micropipette grinder (Narishige) at a 30° angle and a rotor speed of ~ 1800 rpm or 90% of the maximum speed. Needles were backfilled with 3 µL of the RNP mix using Eppendorf Microloader pipette tips. Following injection, the coverslips containing embryos (n = 177 total injected embryos) were placed into 100 X 15 mm Petri dishes containing 1% agarose and held at 28 °C until G0 neonates emerged.
Newly emerged G0 neonates were transferred to individual 30 mL translucent polystyrene cups containing 10 mL of Southland diet and reared at 28 °C (14:10 L:D) to pupation. Pupae (n = 125) were sexed and transferred to individual 30 mL cups until adult eclosion. Two G0 adult females showing full yellow eyes were allowed to mate with two G0 males showing mosaic eyes to generate Yellow-R2 G1 neonates used in diet selection bioassays (below). The remainder of the surviving G0 adults (28 males and 27 females) were placed into cages with 8–10 males and 8–10 females and used to generate Hz-R2 G1 neonates used for selection (below).
Creation of the Cry2Ab-resistant strains Hz-R2 and Yellow-R2
We tested G1 larvae from the unselected Yellow-R2 and Hz-R2 strains for susceptibility to Cry2Ab using our standard 7-d diet overlay bioassays41. We tested one neonate per well in bioassay trays (BIO-BA-128, Pitman, NJ) on diet treated with 1 μg Cry2Ab per cm2 diet (n = 192 for Hz-R2 and n = 224 for Yellow-R2) or on control diet (n = 16) treated with 40 µl of 0.1% Triton X-100 (no Cry2Ab). Trays were covered with Pull N' Peel covers (BIO-CU-16, Pitman, NJ). Survivors (≥ 4th instar larvae at 7 days) on Cry2Ab-treated diet were used to further propagate the Hz-R2 and Yellow-R2 strains. We calculated adjusted survival (%) as the survival (%) on diet containing 1 μg Cry2Ab per cm2 divided by survival (%) on untreated diet. We used a two-tailed Fisher's exact test (http://www.graphpad.com/quickcalcs/contingency1/) to determine if a significant difference occurred between LAB-S and the G1 of each CRISPR-edited strain in the proportion of live larvae on treated diet and on untreated diet.
Cas9-induced mutations in gDNA target regions
To determine if G1 larvae of Yellow-R2 and Hz-R2 that survived on diet surface treated with 1 μg Cry2Ab per cm2 harbored mutations corresponding to sgRNA target sites, we amplified, cloned, and DNA sequenced the relevant HzABCA2 and HzTO gDNA corresponding to sgRNA target sites. We extracted gDNA separately from LAB-S, Yellow-R2, and Hz-R2 larvae using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). Phusion Green High-Fidelity DNA Polymerase (Thermo Fisher Scientific) was used with oligonucleotide primers (Supplementary Table S2) to amplify specific regions corresponding to both HzABCA2 and HzTO sgRNA target sites. PCR conditions were 98 °C for 1 min for 1 cycle; 98 °C for 5 s, 48 °C for 5 s, and 72 °C for 10 s for 35 cycles; 72 °C for 1 min; and hold at 16 °C. PCR amplicons were analyzed on 1.5% agarose gels and stained with SYBR Safe DNA Gel Stain (Thermo Fisher Scientific). DNA bands were excised, cloned into the pJET1.2 vector (Thermo Fisher Scientific), and transformed into One Shot TOP10 Chemically Competent E. coli (Thermo Fisher Scientific). Purified plasmid DNA was Sanger sequenced by Retrogen (San Diego, CA). Multiple sequence alignments were performed using MUSCLE43. The full-length HzABCA2 sequence from LAB-S is deposited in the GenBank public database (Accession number OP186036.1).
Susceptibility to Cry2Ab of Hz-R2 and LAB-S
We used diet overlay bioassays41 to determine the concentration of Cry2Ab necessary to kill 50% of larvae (LC50) for Hz-R2 and LAB-S. Newly emerged neonates (n = 48 larvae per concentration) were placed into separate wells of bioassay trays. For LAB-S these contained 0, 0.1, 0.3, or 1 μg Cry2Ab per cm2 diet, and for Hz-R2 (G4) they contained 0, 1, 3, 10, and 30 μg Cry2Ab per cm2 diet. Trays were held at 28 °C (14 h light:10 h dark). After 7 days, we scored live larvae that were third or higher instars as survivors. We adjusted mortality for control mortality using Abbott’s correction and calculated LC50 using R (v 3.6.3)44 and the publicly available script https://github.com/JuanSilva89/Probit-analysis/commit/2eaaff05da0f89294788bd0bed564e1bf257acf2. The resistance ratio was calculated by dividing the LC50 for Hz-R2 by the LC50 for LAB-S.
Inheritance of resistance to Cry2Ab in strain Hz-R2: maternal effects, sex linkage, and dominance
We evaluated the mode of inheritance of resistance to Cry2Ab for Hz-R2 by testing F1 neonates from reciprocal Hz-R2 X LAB-S crosses in diet overlay bioassays on 1 μg Cry2Ab per cm2 diet (n = 130 neonates per cross) and control diet (n = 32 neonates per cross). Survivors (≥ 4th instar larvae) were determined after 7 days of diet exposure and adjusted % survival was calculated by dividing survival on treated diet by survival on diet without Cry2Ab. We evaluated the dominance parameter h, which varies from 0 for completely recessive resistance to 1 for completely dominant resistance45. We calculated h for Hz-R2 as: h = (F1 survival − S survival)/(R survival − S survival), where the S is LAB-S, R is Hz-R2, and F1 is larvae resulting from reciprocal crosses between Hz-R2 and LAB-S.
Data availability
The full-length HzABCA2 cDNA sequence (OP186036.1) generated during the current study is available in the NCBI GenBank repository (https://www.ncbi.nlm.nih.gov/genbank/).
References
Sanahuja, G., Banakar, R., Twyman, R. M., Capell, T. & Christou, P. Bacillus thuringiensis: A century of research, development and commercial applications. Plant Biotechnol. J. 9, 283–300. https://doi.org/10.1111/j.1467-7652.2011.00595.x (2011).
National Academies of Sciences, E. & Medicine. Genetically Engineered Crops: Experiences and Prospects. (The National Academies Press, Washington, DC, 2016).
Hutchison, W. D. et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 330, 222–225. https://doi.org/10.1126/science.1190242 (2010).
Dively, G. P. et al. Regional pest suppression associated with widespread Bt maize adoption benefits vegetable growers. Proc. Natl. Acad. Sci. USA 115, 3320–3325. https://doi.org/10.1073/pnas.1720692115 (2018).
Romeis, J., Naranjo, S. E., Meissle, M. & Shelton, A. M. Genetically engineered crops help support conservation biological control. Biol. Control 130, 136–154. https://doi.org/10.1016/j.biocontrol.2018.10.001 (2019).
Tabashnik, B. E. et al. Transgenic cotton and sterile insect releases synergize eradication of pink bollworm a century after it invaded the United States. Proc. Natl. Acad. Sci. USA 118, e2019115118. https://doi.org/10.1073/pnas.2019115118 (2021).
Tabashnik, B. E. & Carrière, Y. Global patterns of resistance to Bt crops highlighting pink bollworm in the United States, China, and India. J. Econ. Entomol. 112, 2513–2523. https://doi.org/10.1093/jee/toz173 (2019).
Heckel, D. G. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic. Biochem. Physiol. 104, 103–110. https://doi.org/10.1016/j.pestbp.2012.05.007 (2012).
Tabashnik, B. E. ABCs of insect resistance to Bt. PLoS Genet. 11, e1005646. https://doi.org/10.1371/journal.pgen.1005646 (2015).
Heckel, D. G. The essential and enigmatic role of ABC transporters in Bt resistance of noctuids and other insect pests of agriculture. Insects 12, 389. https://doi.org/10.3390/insects12050389 (2021).
Gahan, L. J., Pauchet, Y., Vogel, H. & Heckel, D. G. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6, e1001248. https://doi.org/10.1371/journal.pgen.1001248 (2010).
Baxter, S. W. et al. Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics 189, 675–679. https://doi.org/10.1534/genetics.111.130971 (2011).
Atsumi, S. et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 109, E1591–E1598. https://doi.org/10.1073/pnas.1120698109 (2012).
Xiao, Y. et al. Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci. Rep. 4, 6184. https://doi.org/10.1038/srep06184 (2014).
Coates, B. S. & Siegfried, B. D. Linkage of an ABCC transporter to a single QTL that controls Ostrinia nubilalis larval resistance to the Bacillus thuringiensis Cry1Fa toxin. Insect Biochem. Mol. Biol. 63, 86–96. https://doi.org/10.1016/j.ibmb.2015.06.003 (2015).
Flagel, L., et al. Mutational disruption of the ABCC2 gene in fall armyworm, Spodoptera frugiperda, confers resistance to the Cry1Fa and Cry1A.105 insecticidal proteins. Sci. Rep. 8, 7255. https://doi.org/10.1038/s41598-018-25491-9 (2018).
Tay, W. T. et al. Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an ABC transporter subfamily A protein. PLoS Genet. 11, e1005534. https://doi.org/10.1371/journal.pgen.1005534 (2015).
Yang, X. et al. Mutation of ABC transporter ABCA2 confers resistance to Bt toxin Cry2Ab in Trichoplusia ni. Insect Biochem. Mol. Biol. 112, 103209. https://doi.org/10.1016/j.ibmb.2019.103209 (2019).
Mathew, L. G. et al. ABC transporter mis-splicing associated with resistance to Bt toxin Cry2Ab in laboratory- and field-selected pink bollworm. Sci. Rep. 8, 13531. https://doi.org/10.1038/s41598-018-31840-5 (2018).
Fabrick, J. A. et al. Shared and independent genetic basis of resistance to Bt toxin Cry2Ab in two strains of pink bollworm. Sci. Rep. 10, 7988. https://doi.org/10.1038/s41598-020-64811-w (2020).
Wang, J. et al. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 87, 147–153. https://doi.org/10.1016/j.ibmb.2017.07.002 (2017).
Li, X. et al. ATP-binding cassette subfamily A member 2 is a functional receptor for Bacillus thuringiensis Cry2A toxins in Bombyx mori, but not for Cry1A, Cry1C, Cry1D, Cry1F, or Cry9A toxins. Toxins 12, 104. https://doi.org/10.3390/toxins12020104 (2020).
Fabrick, J. A. et al. CRISPR-mediated mutations in the ABC transporter gene ABCA2 confer pink bollworm resistance to Bt toxin Cry2Ab. Sci. Rep. 11, 10377. https://doi.org/10.1038/s41598-021-89771-7 (2021).
Luttrell, R. G. & Jackson, R. E. Helicoverpa zea and Bt cotton in the United States. GM Crops Food 3, 1–15. https://doi.org/10.4161/gmcr.20742 (2012).
Reisig, D. D. et al. Long-term empirical and observational evidence of practical Helicoverpa zea resistance to cotton with pyramided Bt toxins. J. Econ. Entomol. 111, 1824–1833. https://doi.org/10.1093/jee/toy106 (2018).
Reisig, D. D. & Kurtz, R. Bt resistance implications for Helicoverpa zea (Lepidoptera: Noctuidae) insecticide resistance management in the United States. Environ. Entomol. 47, 1357–1364. https://doi.org/10.1093/ee/nvy142 (2018).
Bilbo, T. R., Reay-Jones, F. P. F., Reisig, D. D., Greene, J. K. Susceptibility of corn earworm (Lepidoptera: Noctuidae) to Cry1A.105 and Cry2Ab2 in North and South Carolina. J. Econ. Entomol 112, 1845–1857. https://doi.org/10.1093/jee/toz062 (2019).
Kaur, G. et al. Field-evolved resistance of Helicoverpa zea (Boddie) to transgenic maize expressing pyramided Cry1A.105/Cry2Ab2 proteins in northeast Louisiana, the United States. J. Invertebr. Pathol. 163, 11–20. https://doi.org/10.1371/journal.pone.0221343 (2019).
Yang, F., Santiago González, J. C., Williams, J., Cook, D. C., Gilreath, R. T., Kerns, D. L. Occurrence and ear damage of Helicoverpa zea on transgenic Bacillus thuringiensis maize in the field in Texas, U.S. and its susceptibility to Vip3A protein. Toxins 11, 102. https://doi.org/10.3390/toxins11020102 (2019).
Carrière, Y., Degain, B. A., Tabashnik, B. E. Effects of gene flow between Bt and non-Bt plants in a seed mixture of Cry1A.105 + Cry2Ab corn on performance of corn earworm in Arizona. Pest Manag. Sci. 77, 2106–2113. https://doi.org/10.1002/ps.6239 (2021).
Yu, W., et al. Extended investigation of field-evolved resistance of the corn earworm Helicoverpa zea (Lepidoptera: Noctuidae) to Bacillus thuringiensis Cry1A.105 and Cry2Ab2 proteins in the southeastern United States. J. Invertebr. Pathol. 183, 107560. https://doi.org/10.1016/j.jip.2021.107560 (2021).
Yang, F. et al. Practical resistance to Cry toxins and efficacy of Vip3Aa in Bt cotton against Helicoverpa zea. Pest Manag. Sci. https://doi.org/10.1002/ps.7142 (2022).
Yang, F., Santiago González, J. C., Sword, G. A. & Kerns, D. L. Genetic basis of resistance to the Vip3Aa Bt protein in Helicoverpa zea. Pest Manag. Sci. 77, 1530–1535. https://doi.org/10.1002/ps.6176 (2021).
Jin, L. et al. Dominant point mutation in a tetraspanin gene associated with field-evolved resistance of cotton bollworm to transgenic Bt cotton. Proc. Natl. Acad. Sci. USA 115, 11760–11765. https://doi.org/10.1073/pnas.1812138115 (2018).
Fritz, M. L., Nunziata, S. O., Guo, R., Tabashnik, B. E., Carrière, Y. Mutations in a novel cadherin gene associated with Bt resistance in Helicoverpa zea. G3. Genet. 10, 1563–1574. https://doi.org/10.1534/g3.120.401053 (2020).
Benowitz, K. M., et al. Novel genetic basis of resistance to Bt toxin Cry1Ac in Helicoverpa zea. Genetics. 221, iyac037. https://doi.org/10.1093/genetics/iyac037 (2022).
Perera, O. P., Little, N. S. & Pierce, C. A. III. CRISPR/Cas9 mediated high efficiency knockout of the eye color gene Vermillion in Helicoverpa zea (Boddie). PLoS ONE 13, e0197567. https://doi.org/10.1371/journal.pone.0197567 (2018).
Mahon, R. J., Olsen, K. M., Garsia, K. A. & Young, S. R. Resistance to Bacillus thuringiensis toxin Cry2Ab in a strain of Helicoverpa armigera (Lepidoptera: Noctuidae) in Australia. J. Econ. Entomol. 100, 894–902. https://doi.org/10.1093/jee/100.3.894 (2007).
Knight, K. M., Head, G. P. & Rogers, J. D. Successful development and implementation of a practical proactive resistance management plan for Bt cotton in Australia. Pest Manag. Sci. 77, 4262–4273. https://doi.org/10.1002/ps.6490 (2021).
Taylor, K. L., Hamby, K. A., DeYonke, A. M., Gould, F. & Fritz, M. L. Genome evolution in an agricultural pest following adoption of transgenic crops. Proc. Natl. Acad. Sci. USA 118, e2020853118. https://doi.org/10.1073/pnas.2020853118 (2021).
Welch, K. L. et al. Cross-resistance to toxins used in pyramided Bt crops and resistance to Bt sprays in Helicoverpa zea. J. Invertebr. Pathol. 132, 149–156. https://doi.org/10.1016/j.jip.2015.10.003 (2015).
Liu, L. et al. Identification and evaluations of novel insecticidal proteins from plants of the Class Polypodiopsida for crop protection against key lepidopteran pests. Toxins 11, 383. https://doi.org/10.3390/toxins11070383 (2019).
Edgar, R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. https://doi.org/10.1093/nar/gkh340 (2004).
R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, https://www.r-project.org/index.html (2020).
Liu, Y. B. & Tabashnik, B. E. Inheritance of resistance to the Bacillus thuringiensis toxin Cry1C in the diamondback moth. Appl. Environ. Microbiol. 63, 2218–2223. https://doi.org/10.1128/aem.63.6.2218-2223.1997 (1997).
Acknowledgements
We thank Corteva Agriscience for providing Cry2A.127 used in this study. C.H. was supported, in part, by an appointment to the Agricultural Research Service (ARS) Research Participation Program admistered by the Oak Ridge Institute and Science Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by Oak Ridge Associated Universities (ORAU) under DOE contract #DE-SC0014664. All opinions expressed in this paper are the authors' and do not necessarily reflect the policies and views of USDA, DOE, or ORAU/ORISE. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Funding
This article was funded by USDA National Institute of Food and Agriculture, Agriculture and Food Research Initiative (2018-67013-27821).
Author information
Authors and Affiliations
Contributions
J.F, C.H., Y.W., and B.T. designed the study. J.F., C.H., D.L., B.D., A.Y., and G.U. performed the experiments. J.F., C.H., Y.C., and B.T. analyzed the data. J.F., C.H., X.L., Y.C., and B.T. wrote the manuscript. All authors have read and approved the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
This is a cooperative investigation between USDA ARS and the University of Arizona, with J.A.F., Y.C., and B.E.T. receiving funding from USDA National Institute of Food and Agriculture (Agriculture and Food Research Initiative Program Grant #2018-67013-27821) to support this work. C.C.H., D.M.L., B.A.D., A.J.Y., G.C.U, and Y.W., declare no potential conflict of interest. J.A.F. is coauthor of a patent “Cadherin Receptor Peptide for Potentiating Bt Biopesticides” (patent numbers: US20090175974A1, US8354371, WO2009067487A2, WO2009067487A3). B.E.T. is a coauthor of a patent on modified Bt toxins, "Suppression of Resistance in Insects to Bacillus thuringiensis Cry Toxins, Using Toxins that do not Require the Cadherin Receptor" (patent numbers: CA2690188A1, CN101730712A, EP2184293A2, EP2184293A4, EP2184293B1, WO2008150150A2, WO2008150150A3). Bayer Crop Science, BASF, Corteva Agriscience, and Syngenta did not provide funding to support this work but may be affected financially by publication of this paper and have funded other work by J.A.F., Y.C., X.L., and/or B.E.T.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fabrick, J.A., Heu, C.C., LeRoy, D.M. et al. Knockout of ABC transporter gene ABCA2 confers resistance to Bt toxin Cry2Ab in Helicoverpa zea. Sci Rep 12, 16706 (2022). https://doi.org/10.1038/s41598-022-21061-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-21061-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.