Abstract
We investigated the association between genetic variants in the histone modification regions and the prognosis of lung adenocarcinoma after curative surgery. Potentially functional SNPs were selected using integrated analysis of ChIP-seq and RNA-seq. The SNPs were analyzed in a discovery set (n = 166) and a validation set (n = 238). The associations of the SNPs with overall survival (OS) and disease-free survival (DFS) were analyzed. A total of 279 SNPs were selected for genotyping. Among these, CAPN1 rs17583C>T was significantly associated with better OS and DFS (P = 0.001 and P = 0.007, respectively), and LINC00959 rs4751162A>G was significantly associated with worse DFS (P = 0.008). Luciferase assays showed a significantly lower promoter activity of CAPN1 in the rs17583 T allele than C allele (P = 0.008), and consistently the CT + TT genotypes had significantly lower CAPN1 expression than CC genotype (P = 0.01) in clinical samples. The rs4751162 G allele had higher promoter activity of GLRX3 than A allele (P = 0.05). The motif analyses and ChIP-qPCR confirmed that the variants are located in the active promoter/enhancer regions where transcription factor binding occurs. This study showed that genetic variants in the histone modification regions could predict the prognosis of lung adenocarcinoma after surgery.
Similar content being viewed by others
Introduction
Carcinogenic process can be driven by the accumulation of epigenetic alterations as well as genetic alterations1. Epigenetics comprise heritable modifications to the chromatin that affect gene expression without altering DNA coding sequence, including DNA methylation, microRNA regulation, and histone modifications2. These epigenetic changes and their roles in human cancer have been actively studied for the past decades2. The knowledge of various epigenetic events has led to deeper understanding of the pathogenesis of cancer, and provided a clue to the discovery of prognostic biomarkers and novel therapeutic targets1. Histones are the central component of the nucleosomes, the fundamental building blocks of chromatin. The histone tails are subject to extensive posttranslational modifications such as methylation and acetylation among others, which can contribute to chromatin compaction, nucleosome dynamics, and transcriptional processes3. Dysregulation of these processes may lead to aberrant gene expression, which is frequently observed in human cancers4. Many studies have reported the role of genetic variations in the regulation of epigenome5,6,7,8, and their association with the risk and clinical outcomes of human cancers9,10,11,12.
Lung cancer is one of the most commonly diagnosed malignancies and is the leading cause of cancer-related death worldwide, with an average 5-year survival rate of 19%13. Lung adenocarcinoma comprises approximately 50% of all lung cancer14. Advanced understanding in molecular pathogenesis have enabled the identification of driver mutations and the development of personalized treatment options with clinically meaningful outcomes in metastatic lung adenocarcinoma15,16. In early-stage lung adenocarcinoma, many patients experience recurrence and even death after complete resection although surgery is the best treatment modality for cure. Even patients with the same stage are regarded to be at a variable risk of recurrence. However, reliable prognostic biomarkers are still not available. Therefore, deeper understanding in the molecular pathogenesis is essential to promote the development of prognostic biomarkers for building effective post-surgical strategies such as adjuvant therapy and follow-up to improve prognosis in early-stage NSCLC.
In this study, we hypothesized that potentially functional single nucleotide polymorphisms (SNPs) in histone modification regions may modulate the pathogenesis of lung cancer by regulating the expression of target genes, and consequently clinical outcomes of lung cancer. To test this hypothesis, we aimed to investigate the association between functional variants within or adjacent to genes with high expression in histone modification regions and the prognosis after surgery in lung adenocarcinoma.
Results
Patient characteristics and clinical outcomes
The baseline clinical and pathologic characteristics of patients in the discovery and validation cohorts and their association with OS and DFS are shown in Table 1. Median duration of follow-up was 30.4 (range, 3.6–67.1) months for the discovery cohort, and 27.4 (range, 1.0–66.7) months for the validation cohort. Pathologic stage was significantly associated with OS and DFS in both cohorts (log-rank P [PL–R] for OS = 0.01 and 3 × 10–6, and PL–R for DFS = 4 × 10–5 and 3 × 10–10 in the discovery and the validation cohorts, respectively). In the discovery cohort, age was significantly associated with OS (PL–R = 0.02), and adjuvant therapy was associated with DFS (PL–R = 0.04). In the validation cohort, sex and smoking status were significantly associated with OS and DFS (PL–R for OS = 0.04 and 0.01; and PL–R for DFS = 0.03 and 0.01, respectively).
Identification of potentially functional SNPs using ChIP-seq and RNA-seq
The regulatory variants in histone modification regions may affect the expression of genes. To identify potentially functional variants in histone modification regions, we performed ChIP-seq with antibodies against two posttranslational modifications of histone H3 (H3K4me3 and H3K27ac) and RNA-seq using H2087 cell lines. Based on the ChIP-seq data for two histone marks, 31,582 SNPs located within H3K4me3 peaks and 34,591 within H3K27ac peaks were retrieved. And then, based on the NCBI SNP database (https://snpinfo.niehs.nih.gov/), 1654 SNPs within H3K4me3 peaks and 1229 within H3K27ac peaks, which were potentially functional and had minor allele frequency ≥ 0.1, were collected. Using RNA-seq we chose genes with high expression level (FPKM ≥ 100), and then 383 SNPs within or closest to the genes were extracted, of which 320 were identified as tag SNPs. Finally, the 279 SNPs were selected because 41 located within the overlap between the H3K4me3 peaks and H3K27ac peaks were duplicated.
Associations between the SNPs and survival outcomes
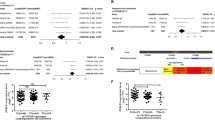
Among the 279 SNPs genotyped, 184 were analyzed for the association study after excluding 10 with genotyping failure, 85 with deviation from the HWE (P < 0.05) or call rate < 90%. In the discovery set, 41 SNPs were significantly associated with clinical outcomes (Supplementary Table S1). Among 41 SNPs, two (CAPN1 rs17583C>T and LINC00959 rs4751162A>G) were found to be significantly associated with survival outcomes in a validation set (Supplementary Table S2). In a combined analysis that adjusted for age, gender, smoking status, pathologic stage, and adjuvant therapy, CAPN1 rs17583C>T was significantly associated with better OS and DFS (adjusted HR [aHR] = 0.49, 95% CI 0.32–0.75, P = 0.001, under a dominant model; aHR = 0.43, 95% CI 0.23–0.79, P = 0.007, under a recessive model, respectively), and LINC00959 rs4751162A>G exhibited worse DFS (aHR = 1.53, 95% CI 1.11–2.09, P = 0.008, under a dominant model) (Fig. 1 and Table 2).
Effect of rs17583 C>T and rs4751162 A>G on promoter activity and mRNA expression
The rs17583C>T is located in the promoter region of CAPN1, and the rs4751162A>G is located in the intron of LINC00959. The rs4751162 resides in an enhancer which is 26 kb apart from GLRX3 and expected to regulate its expression. To verify the functional relevance of the two genetic variants, we investigated whether rs17583C>T and rs4751162A>G regulate the promoter activity of the CAPN1 and GLRX3 gene, respectively. The promoter assays showed that the CAPN1 rs17583 T allele had significantly lower promoter activity than the C allele (Fig. 2A, P = 0.008). The LINC00959 rs4751162 G allele had significantly higher GLRX3 promoter activity than the A allele (Fig. 2B, P = 0.05). Quantitative RT-PCR showed that the relative CAPN1 expression was significantly higher in tumor tissue than in normal lung tissue (Fig. 3A, P = 0.003). In tumor tissues, CAPN1 expression was significantly lower in CT + TT than in CC genotype (Fig. 3B, P = 0.01). The expression of GLRX3 was also significantly higher in tumor tissue than in normal tissue (Fig. 3C, P = 0.0003). GLRX3 expression was not significantly different among genotypes (Fig. 3D). There was no significant association between mRNA expression level and the survival outcomes (data not shown).
The transcription activity according to alleles of CAPN1 rs17583C>T and LINC00959 rs4751162A>G measured by luciferase reporter assay. The H1703 cells were transfected with pGL3-CAPN1_C and pGL3-CAPN1_T for rs17583 (A), and pGL3-GLRX3pro, pGL3-GLRX3pro_A, or pGL3-GLRX3pro_G for rs4751162 (B), respectively. The data were analyzed using Excel. Each bar represents mean ± SEM of luciferase activity normalized to Renilla luciferase activity. Experiments were performed in triplicate. P value, Student's t-test.
The mRNA expression levels of CAPN1 and GLRX3 in tumor and corresponding non-malignant lung tissues (A, n = 73 and C, n = 55), and CAPN1 and GLRX3 mRNA expression according to CAPN1 rs17583C>T (32CC, 29CT, and 8TT) and LINC00959 rs4751162A>G (32AA, 15AG, and 1GG) genotypes (B, D). The data were analyzed using SPSS. The P-value was calculated using Student’s t-test.
ChIP-qPCR assays to confirm histone modification and transcription factor binding in two SNP-containing regions
To confirm whether the two genetic variants are located in functional promoter or enhancer with active histones, we performed ChIP-qPCR assays using antibodies against H3K4me3, H3K27ac, H3K9ac, and H3Kme1. The rs17583-containing region was associated with strong enrichment of H3K4me3 and H3K9ac which mark active promoters (Fig. 4A), and the rs4751162-containing region showed strong enrichment of H3K27ac and H3Kme1 which mark active enhancers (Fig. 4B). Next, motif analyses were performed to predict transcription factors which bind to the SNP regions. As a result of aligning with known motifs using TOMTOM tool in the MEME Suite web server17, the rs17583 and rs4751162 are located in the binding motifs of YY1 and TFAP4, respectively (Supplementary Fig. S1). We performed ChIP assays with antibodies to YY1, TFAP4 or with IgG as a control. The rs17583 and rs4751162 regions showed significant enrichment of YY1 and TFAP4, respectively (Fig. 4C,D). The YY1 was predicted to bind to rs17583 C allele more efficiently than to T allele, and TFAP4 was predicted to bind to rs4751162 G allele more efficiently than to A allele, suggesting the alleles with increased gene expression had preferred transcription factor binding (Supplementary Fig. S1).
ChIP-qPCR analysis of H3K4me3 and H3K9ac (A) and YY1 (C) binding at CAPN1 rs17583, and H3K27ac and H3K4me1 (B) and TFAP4 (D) binding at LINC00959 rs4751162. The data were analyzed using Excel. Each bar represents mean ± SEM from three independent experiments carried out in triplicate. P values by Student’s t-test.
Discussion
In this study, we investigated the association between genetic variation in histone modification regions and the prognosis in lung adenocarcinoma. We found that two genetic variants, CAPN1 rs17583C>T and LINC00959 rs4751162A>G, in the histone modification regions were associated with the survival outcomes of patients with lung adenocarcinoma who underwent curative surgery. Functional analyses showed significant difference in the promoter activity and expression level of CAPN1 and GLRX3 genes according to genotypes, supporting the association between the variants and the survival outcomes. The motif analyses and ChIP-qPCR confirmed that the variants are located in the active promoter/enhancer regions where transcription factor binding occurs, elucidating epigenetic regulatory mechanisms of gene expression by the variants. These results suggested a potential role of the two genetic variants in the pathogenesis of lung adenocarcinoma, and their usefulness as prognostic biomarkers.
Calpains (CAPNs) are a family of calcium-dependent cysteine proteinases involved in a variety of cellular processes including remodeling of cytoskeletal/membrane attachments, signal transduction, and apoptosis18. CAPN1 and CAPN2 are the two isoforms of the ubiquitous calpain, which are heterodimers of a large catalytic subunit encoded by CAPN1 and CAPN2, respectively, and a regulatory subunit (CAPN4) encoded by CAPNS119. CAPNs could cause limited cleavage or functional modulation of various substrates that act as metastatic mediators, and several studies have suggested that it plays a significant role in tumor migration and invasion20. Only a few studies have addressed the role of CAPNs in lung cancer. It has been reported that CAPN2 might promote lung cancer progression by activating EGFR/pAKT signaling pathway, and also contribute to the resistance to paclitaxel or EGFR-TKI21,22. Another study suggested that overexpression of CAPN4 was an independent prognostic factor in patients with NSCLC, and could enhance the invasive potential of lung cancer cells by upregulating the expression of matrix metalloproteinase 223. In the present study, the CAPN1 rs17583C>T was associated with significantly better clinical outcomes. The CAPN1 expression was significantly higher in lung adenocarcinoma than normal lung. Functional analyses showed that rs17583C-to-T change was associated with decreased CAPN1 mRNA expression in clinical samples and significantly decreased CAPN1 promoter activity in in vitro promoter assays. The ChIP-qPCR confirmed that CAPN1 rs17583C>T is located in an active promoter with YY1 binding. These results suggested a tumor promoting function of CAPN1 in lung adenocarcinoma, and provided evidence of the functional relationship between the genetic variant and the better clinical outcomes.
GLRX3 is an antioxidant enzyme, one of the intracellular redox-regulating molecules that contribute to maintaining cellular redox homeostasis, and is known to play an important role in cellular signal transduction in response to stress signals by reactive oxygen species24. Although the complicated roles of GLRX3 in cancer remain poorly understood, overexpression of GLRX3 was ascertained in several types of malignancy, such as nasopharyngeal carcinoma (NPC), oral squamous cell carcinoma (OSCC), colon cancer, and lung cancer25,26,27. Studies on NPC and OSCC have reported in common that knockdown of GLRX3 inhibited cell proliferation and decreased the migration and invasion capacity of cancer cells by suppressing the epithelial-mesenchymal transition, indicating the essential roles of GLRX3 in cancer progression25,26. In agreement with these results, our unpublished data indicated that knockdown of GLRX3 inhibited proliferation, migration, and invasion capacity of lung cancer cells. In this study, LINC00959 rs4751162A>G was associated with worse DFS. GLRX3 expression was significantly higher in tumor tissues than in normal lung, and rs4751162 G allele correlated with a significantly higher promoter activity of GLRX3 than the A allele. Motif analyses predicted that the rs4751162 is located in the binding motif of TFAP4, and the ChIP-qPCR confirmed the SNP was located in an active enhancer with TFAP4 binding. These results suggested a potentially oncogenic role of GLRX3. However, further investigation is needed to understand the role of CAPN1 and GLRX3 in the pathogenesis of lung adenocarcinoma and the biological mechanisms of the association between the genetic variants and clinical outcomes.
In this study, only two out of 41 SNPs were replicated in the validation cohorts. Difference in clinical characteristics of the patients between the discovery and validation cohorts, which could be a possible cause of replication failure, is a limitation of the retrospective multicenter studies. However, all the clinical variables were adjusted for in the multivariate analyses. More importantly, because the discovery study with a relatively small sample size was an exploratory study for which type II errors should be considered, we did not perform multiple testing corrections for the associations, which may have resulted in false positive associations leading to the replication failure of the associations in the validation study. In addition, the modest sample size of the validation cohort may not have optimal statistical power for replicating the associations. In this study, the observed P values did not reach a more stringent level of statistical significance to avoid false positive associations arising from multiple comparisons. Future studies with larger number of patients are required to validate our results. Nevertheless, the design of two-stage independent cohorts for the discovery and validation sets is a major strength of this study, which could reduce false-positive findings from the genetic association study28,29. The two SNPs were significantly associated with clinical outcomes in both independent cohorts, and the association had similar effect size with the same direction. In addition, the association showed higher level of significance in the combined analysis including larger population, supporting the credibility of the association. Functional analyses further supported the plausibility of our findings.
In summary, the current study demonstrated that the two genetic variants, CAPN1 rs17583C>T and LINC00959 rs4751162A>G, was associated with survival outcomes of patients with lung adenocarcinoma after surgical resection. Our results suggest that CAPN1 and GLRX3 may play important roles in the pathogenesis of lung adenocarcinoma, and that the variants may be useful in predicting the prognosis of lung adenocarcinoma after surgery, thereby helping to refine therapeutic decisions for better clinical outcomes in NSCLC. Future studies are warranted to validate our results in a larger population with diverse ethnicity.
Methods
Study population
A total of 404 patients with available genomic DNA samples, who were diagnosed with pathologic stages I, II, or IIIA (micro-invasive N2) NSCLC after curative surgical resection, were enrolled in this study. Discovery cohort comprised 166 patients whose diagnosis was made at Kyungpook National University Hospital (KNUH) between September 2001 and August 2009, and validation cohort consisted of 238 patients diagnosed with NSCLC at Seoul National University Bundang Hospital (SNUBH) between June 2005 and May 2012. All patients in this study were of Korean ethnicity. Genomic DNA samples extracted from peripheral blood lymphocytes of the patients were provided by the National Biobank of Korea, KNUH, which is supported by the Ministry of Health, Welfare and Family Affairs. Written informed consent was obtained from all patients before surgery. This study was approved by the Institutional Review Boards of the Kyungpook National University Chilgok Hospital and Seoul National University Bundang Hospital (Approval No. KNUCH 2017-07-012). All experiments were performed in accordance with relevant guidelines and regulations.
Cell culture and antibodies
H2087 cells were obtained from the American Type Culture Collection (ATCC) and H1703 cells were purchased from Korean Cell Line Bank (KCLB), Seoul, Korea. Antibodies used in this study include anti-Histone H3 antibodies (ab8580, ab4441, ab4729 and ab8895), and anti-GLRX3 antibody (ab226396) from Abcam (Cambridge, UK), anti-YY1 antibody (46395) from Cell Signaling Technology (Danvers, MA, USA), and anti-TFAP4 antibody (sc-166216X) from Santa Cruz Biotechnology (Dallas, TX, USA). Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 in Corning RPMI medium (Corning Inc., Corning, NY, USA) supplemented with 10% Corning Fetal Bovine Serum (Corning Inc., Corning, NY, USA), and 100 U/ml penicillin and 100 mg/ml streptomycin.
Chromatin immunoprecipitation (ChIP)-sequencing
ChIP assays were performed using the Pierce Magnetic ChIP kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s protocol. H2087 cells were crosslinked with 1% formaldehyde for 10 min, and the crosslinking was inactivated by 0.125 M glycine for 5 min at room temperature. Cells were washed with cold 1xPBS twice. The cells were lysed, sonicated to shear DNA. To immunoprecipitate protein/chromatin complexes, the diluted supernatants were incubated with 10 μg of H3K4me3 or H3K27ac antibody overnight, and then incubated for 2 h after adding 50 μl of agarose/protein A or G beads. Ten percent of the diluted supernatants were saved as “input” for normalization. Several washing steps were followed by protein digestion using proteinase K. Reverse crosslinking was carried out at 65 °C. DNA was subsequently purified. ChIPSeq library preparation was performed with TruSeq ChIP Library Preparation Kit (Illumina, San Diego, CA, USA). Sequencing was performed on an Illumina HiSeq4000. Sequence reads for each sample were aligned to the human genome using Bowtie30. The reference genome sequence of Homo sapiens (hg19) and annotation data were downloaded from the UCSC table browser (http://genome.uscs.edu). Peaks were called in the aligned sequence data using a model-based analysis of ChIP-seq (MACS2 version 2.1.0) (https://bioweb.pasteur.fr/packages/pack@macs@2.1.0)31. ChIPseeker (version 1.6.6) (http://www.bioconductor.org/packages/release/bioc/html/ChIPseeker.html)32, a bioconductor package within the statistical programming environment R to facilitate batch annotation of enriched peaks identified from ChIP-seq data, was used to identify nearby genes and transcripts from the peaks obtained from MACS2.
RNA-sequencing
Total RNAs from H2087 cells were isolated using TRIzol (Invitrogen, Carlsbad, CA, USA). Sequencing was performed on an Illumina HiSeq4000 and aligned the processed reads to the Homo sapiens (hg19) using HISAT v2.0.533. Transcript assembly and abundance estimation was performed using StringTie v1.3.3b34,35. It provides the relative abundance estimates as FPKM values (Fragments Per Kilobase of exon per Million fragments mapped) of transcript and gene expressed in each sample. FPKM values have already been normalized with respect to library size.
SNP selection and genotyping
We conducted an integrated analysis of ChIP-seq and RNA-seq for the SNP selection. As a result of the ChIP-seq using H2087 cells, SNPs within H3K4me3 and H3K27ac peak regions were selected. Next, using the FuncPred utility for functional SNP prediction in the SNPinfo web server (https://snpinfo.niehs.nih.gov/), potentially functional variants with minor allele frequency ≥ 0.1 based on the HapMap JPT data were collected after excluding those in linkage disequilibrium (r2 ≥ 0.8). And then, using RNA-seq we chose genes with high expression level (FPKM ≥ 100), and then SNPs within or closest to the genes were extracted. Genotyping was performed using iPLEX Assay and MassARRAY System (Agena Bioscience, San Diego, CA, USA). Approximately 5% of the samples were randomly selected to be genotyped again by a different investigator, by a restriction fragment length polymorphism assay, and the results were 100% concordant.
Promoter-luciferase constructs and luciferase assay
We evaluated the effect of the rs17583C>T or rs4751162A>G on the activity of the promoter of CAPN1 or GLRX3 genes by luciferase reporter assay. The rs17583C>T of CAPN1 gene is located in the region of H3K4me3 peak, which marks active promoters, in CAPN1 gene promoter. The 378 bp fragment including rs17583C>T was synthesized by polymerase chain reaction from human genomic DNA and cloned into XhoI/HindIII site of the pGL3-basic vector (Promega, Madison, WI, USA). The correct sequences of all clones were verified by DNA sequencing. The rs4751162A>G is located in the region of H3K27ac peak, which is an activation mark of enhancers, in the intron region of LINC00959 gene. The SNP is expected to regulate expression of GLRX3 gene because it resides 26 kb downstream of GLRX3 gene, although they both are on the chromosome 10. The promoter region of GLRX3 (− 980 to + 38 bp, the transcriptional start site is designated as + 1) was synthesized by polymerase chain reaction from human genomic DNA and cloned into XhoI/NcoI site of the pGL3-promoter vector (Promega, Madison, WI, USA) to generate pGL3-GLRX3pro. Two fragments including rs4751162A or rs4751162G allele of rs4751162A>G were amplified from genomic DNA sample and the 283 bp products were cloned into BamHI/SalI site of the pGL3-GLRX3pro, respectively, to generate pGL3-GLRX3pro_A and pGL3-GLRX3pro_G. The cloning PCR primers were listed Supplementary Table S3. All constructs were verified by direct sequencing before use. The H1703 cells were transfected with 200 ng of each plasmid DNA (pGL3-CAPN1_C and pGL3-CAPN1_T for rs17583, and pGL3-GLRX3pro, pGL3-GLRX3pro_A, or pGL3-GLRX3pro_G for rs4751162) and 2 ng of pRL-SV40 Vector (Promega, Madison, WI, USA) using Effectene transfection reagent (Qiagen, Hilden, Germany) according to manufacturer’s protocol. The cells were collected 48 h after transfection. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase activity measurements were normalized with respect to pRL-SV40 Renilla luciferase activity to correct for variations in transfection efficiency. All experiments were performed in triplicate.
RNA preparation and quantitative reverse transcription-PCR (qRT-PCR)
CAPN1 and GLRX3 mRNA expression was examined by qRT-PCR. Total RNAs from tumors and paired non-malignant lung tissues (n = 73) were isolated using TRIzol (Invitrogen, Carlsbad, CA, USA). Real-time PCR was performed using a LightCycler 480 (Roche Applied GLRX3 expression Science, Mannheim, Germany) with QuantiFast SYBR Green PCR Master Mix (Qiagen, Hilden, Germany). The real-time PCR primers for CAPN1, GLRX3 and β-actin genes were listed in Supplementary Table S3. Each sample was run in duplicate. Relative target gene mRNA expression was normalized to that of β-actin expression and then evaluated using the 2−ΔΔCt method36.
ChIP-quantitative PCR (qPCR) assay
Chromatin from H2087 cells was immunoprecipitated with the Pierce Magnetic ChIP kit (Thermo Fisher Scientific, Waltham, MA, USA), using 10 μg anti-H3K4me3, anti-H3K9ac, anti-H3K27ac, anti-H3Kme1, anti-YY1, and anti-TFAP4 antibodies and 2 μg normal rabbit IgG antibodies per immunoprecipitation reaction. Immunoprecipitated chromatin was subjected to real-time qPCR using SYBR Green PCR Master Mix (Qiagen, Hilden, Germany). The ChIP-qPCR primers were listed Supplementary Table S3. The qPCR was performed as follows: 95 ℃ 10 min, 45 cycles of 95 ℃ 15 s, 60 ℃ 1 min. ChIP-qPCR enrichment analysis were performed by Comparative Ct method. Each samples were normalized to the input and the fold difference between sample and IgG was calculated using 2(− ΔΔCt)37.
Enriched motif analysis
We identified the transcription factor binding motif enriched in the regions containing rs17583C>T or rs4751162 A>G. Motifs were analyzed using TOMTOM, a motif database scanning algorithm, of the MEME Suite web server17 for comparison against Human and Mouse38 and the SwissRegulon databases of known transcription factor motifs39.
Statistical analyses
Hardy–Weinberg equilibrium was evaluated by a goodness-of-fit χ2 test with 1 degree of freedom. Overall survival (OS) was measured from the date of surgery to the date of death or the last follow-up. Disease-free survival (DFS) was calculated from the date of surgery until first evidence of disease recurrence or last date of follow up for patients who were free of disease. Estimated survival rate was calculated using the Kaplan–Meier method. Log-rank test was used to compare the difference in OS and DFS across different genotypes. Multivariate Cox proportional hazards models were used to estimate the hazard ratio (HR) and 95% confidence intervals (CI) after adjusting for age (< 64 years vs ≥ 64 years), gender (male vs female), smoking status (never vs ever), pathological stage (I vs II–IIIA), and adjuvant therapy (yes vs no). Statistical analyses were carried out using Statistical Analysis System for Windows, version 9.4 (SAS Institute, Cary, NC, USA), Statistical Package for the Social Sciences (SPSS) 25.0 (IBM Corp., Armonk, NY, USA), and Microsoft Excel (Microsoft Corp., Redmond, WA, USA).
Data availability
The datasets for the RNA-seq and ChIP-seq (H3K4me3, H3K27ac) have been deposited and are available at Gene Expression Omnibus (GEO accession no. GSE182385) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182385).
References
Langevin, S. M., Kratzke, R. A. & Kelsey, K. T. Epigenetics of lung cancer. Transl. Res. 165, 74–90 (2015).
Ansari, J., Shackelford, R. E. & El-Osta, H. Epigenetics in non-small cell lung cancer: From basics to therapeutics. Transl. Lung Cancer Res. 5, 155 (2016).
Audia, J. E. & Campbell, R. M. Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8, a019521 (2016).
Zhao, Z. & Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 20, 245 (2019).
Heinz, S. et al. Effect of natural genetic variation on enhancer selection and function. Nature 503, 487–492 (2013).
McDaniell, R. et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328, 235–239 (2010).
McVicker, G. et al. Identification of genetic variants that affect histone modifications in human cells. Science 342, 747–749 (2013).
Banovich, N. E. et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 10, e1004663 (2014).
Hong, M. J. et al. A genetic variation in microRNA target site of ETS2 is associated with clinical outcomes of paclitaxel-cisplatin chemotherapy in non-small cell lung cancer. Oncotarget 7, 15948–15958 (2016).
Huang, Q. et al. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet. 46, 126–135 (2014).
Zhu, J. et al. Polymorphism of DNA methyltransferase 3B-149C/T and cancer risk: A meta-analysis. Med. Oncol. 32, 399 (2015).
Tang, D. et al. Novel genetic variants in HDAC2 and PPARGC1A of the CREB-binding protein pathway predict survival of non-small-cell lung cancer. Mol. Carcinog. 59, 104–115 (2020).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Meza, R., Meernik, C., Jeon, J. & Cote, M. L. Lung cancer incidence trends by gender, race and histology in the United States, 1973–2010. PLoS ONE 10, e0121323 (2015).
Mok, T. S. Personalized medicine in lung cancer: What we need to know. Nat. Rev. Clin. Oncol. 8, 661–668 (2011).
Korpanty, G. J., Graham, D. M., Vincent, M. D. & Leighl, N. B. Biomarkers that currently affect clinical practice in lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front. Oncol. 4, 204 (2014).
Bailey, T. L. et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue), W202–W208 (2009).
Goll, D. E., Thompson, V. F., Li, H., Wei, W. & Cong, J. The calpain system. Physiol. Rev. 83, 731–801 (2003).
Suzuki, K., Sorimachi, H., Yoshizawa, T., Kinbara, K. & Ishiura, S. Calpain: Novel family members, activation, and physiological function. Biol. Chem. Hoppe Seyler 376, 523–529 (1995).
Chen, J., Wu, Y., Zhang, L., Fang, X. & Hu, X. Evidence for calpains in cancer metastasis. J. Cell. Physiol. 234, 8233–8240 (2019).
Xu, F. et al. Calpain-2 enhances non-small cell lung cancer progression and chemoresistance to paclitaxel via EGFR-pAKT pathway. Int. J. Biol. Sci. 15, 127–137 (2019).
Zhang, G. et al. Calpain 2 knockdown promotes cell apoptosis and restores gefitinib sensitivity through epidermal growth factor receptor/protein kinase B/survivin signaling. Oncol. Rep. 40, 1937–1946 (2018).
Gu, J. et al. Capn4 promotes non-small cell lung cancer progression via upregulation of matrix metalloproteinase 2. Med. Oncol. 32, 51 (2015).
Lillig, C. H., Berndt, C. & Holmgren, A. Glutaredoxin systems. Biochim. Biophys. Acta. 1780, 1304–1317 (2008).
Li, B. et al. Glutaredoxin 3 promotes migration and invasion via the Notch signalling pathway in oral squamous cell carcinoma. Free Radic. Res. 52, 390–401 (2018).
He, F. et al. Glutaredoxin 3 promotes nasopharyngeal carcinoma growth and metastasis via EGFR/Akt pathway and independent of ROS. Oncotarget 7, 37000–37012 (2016).
Cha, M.-K. & Kim, I.-H. Preferential overexpression of glutaredoxin3 in human colon and lung carcinoma. Cancer Epidemiol. 33, 281–287 (2009).
Pharoah, P. D., Dunning, A. M., Ponder, B. A. & Easton, D. F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 4, 850–860 (2004).
Chanock, S. J. et al. Replicating genotype–phenotype associations. Nature 447, 655–660 (2007).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Yu, G., Wang, L.-G. & He, Q.-Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods 25, 402–408 (2001).
Mukhopadhyay, A., Deplancke, B., Walhout, A. J. & Tissenbaum, H. A. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3, 698–709 (2008).
Jolma, A. et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013).
Pachkov, M., Balwierz, P. J., Arnold, P., Ozonov, E. & Nimwegen, E. V. SwissRegulon, a database of genome-wide annotations of regulatory sites: Recent updates. Nucleic Acids Res. 41(Database issue), D214–D220 (2013).
Acknowledgements
This study was supported by the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (Grant number: 1720040), and by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (No. NRF-2020R1A5A2017323).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: S.Y.L., and J.Y.P. Performed the experiments: H.-G.K., J.E.C., S.K.D., M.J.H., and J.H.L. Acquired clinical data: Y.H.L., S.Y.L., J.Y.J., Y.W.D., E.B.L., K.M.S., S.H.C., H.S., S.S.Y., J.L., S.I.C., C.H.K., S.C., S.J., and J.Y.P. Analyzed and interpreted the data: H.-G.K., Y.H.L., W.K.L., S.Y.L., and J.Y.P. Wrote the main manuscript text: H.-G.K., Y.H.L., S.Y.L., and J.Y.P. Supervised the study: S.Y.L. and J.Y.P. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kang, HG., Lee, Y.H., Lee, S.Y. et al. Genetic variants in histone modification regions are associated with the prognosis of lung adenocarcinoma. Sci Rep 11, 21520 (2021). https://doi.org/10.1038/s41598-021-00909-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-00909-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.