Abstract
Insect midgut microbial symbionts have been considered as an integral component in thermal adaptation due to their differential thermal sensitivity. Altered midgut microbial communities can influence both insect physiology and competence for important vector-borne pathogens. This study sought to gain insights into how Aedes aegypti midgut microbes and life history traits are affected by increase in baseline diurnal temperature. Increase in temperature resulted in the enrichment of specific taxa with Bacillus being the most enriched. Bacillus is known to be heat tolerant. It also resulted in a dissimilar microbial assemblage (Bray–Curtis Index, PERMANOVA, F = 2.2063; R2 = 0.16706; P = 0.002) and reduced survivorship (Log-rank [Mantel-Cox] test, Chi-square = 35.66 df = 5, P < 0.0001). Blood meal intake resulted in proliferation of pathogenic bacteria such as Elizabethkingia in the midgut of the mosquitoes. These results suggest that alteration of temperature within realistic parameters such as 2 °C for Ae. aegypti in nature may impact the midgut microbiome favoring specific taxa that could alter mosquito fitness, adaptation and vector–pathogen interactions.
Similar content being viewed by others
Introduction
The global mean surface temperature is predicted to rise by 1.6–6.6 °C in the late twenty-first Century1. Physiology2,3,4, behavior5, ecology6,7 and evolutionary history8,9,10 may influence the biological responses of organisms to global climate change. Aedes aegypti is the vector of both Zika and dengue viruses which are of global health importance. Dengue fever has spread to five continents resulting in over 3 billion people at risk of contracting the disease11 while emergent Zika outbreaks have resulted in a public health emergencies of international concern.
Recently, it has become clear that microbial symbionts often facilitate or limit adaptation to environmental fluctuations12. Hence, species and communities’ response to climate change may be significantly impacted by the adaptability of their microbial partners. Insect midguts provide a receptive environment for microbial colonization and the midgut bacteria play a myriad of important roles in the survival of their host13,14,15. Midgut bacteria of insects have demonstrated properties including: nutrition, protection from parasites and pathogens, modulation of immune responses, mating and communication16.
It is therefore important to understand how the microbes of medically important vectors such as Aedes mosquitoes will react to increase in temperature as this may provide insights into how this vector will change in response to climate variation. By extension, this will allow the projection into how the landscape of the diseases of global importance caused by the pathogens they transmit will be altered.
The impact of temperature change on the midgut microbiome diversity of insects has mainly been studied with constant temperature. The results of such studies show a significant reduction in the richness of the midgut microbiome to complete loss of midgut microbiome as well as reduction in the vertical transmission to the next generation17,18,19,20. Given that temperatures fluctuate in nature; using constant temperature to study the impact of temperature on the midgut microbiome may not provide an accurate depiction. This gap in knowledge is highly important given the well documented role of the midgut microbiome in vectorial capacity21,22,23,24,25,26,27,28. By adjusting the baseline temperature to the future predicted increase in temperature, we studied the life history traits and midgut microbiome of Ae. aegypti from two geographical locations. We tested the hypothesis that significant variation in composition of Ae. aegypti midgut microbiome is associated with increased temperature. The aims of this study were: to delineate the impact of increase in diurnal temperature on midgut microbiome composition and mosquito life history traits; to describe the effect of non-infectious blood meal intake on the midgut microbial diversity and finally to examine population differences in microbial assemblage, comparing two Ae. aegypti populations.
Results
Temperature increase impacts the immature stages and adult emergence of Aedes in a population-specific manner
The immature stages of the mosquitoes were reared in three replicates which were statistically similar, hence all the calculations on the impact of temperature on the immature stages as well as adult emergence were based on mean values of the biological replicates. Increase in temperature had a population-specific impact on the egg eclosion rates and larval death rates. The egg eclosion rates of the Miami population improved with an increase in temperature (Fisher’s exact test P < 0.0001) (Table 1); the converse was observed in the Mexico population as shown by a reduction in their egg eclosion rates when the temperature was increased (Fisher’s exact test P < 0.0001) (Table 1). Between the two populations at the L temperature regime, AEG MX had a higher egg eclosion rate than AEG MI (Chi-square = 31.52, df = 1, P < 0.0001) (Table 1). In contrast, egg eclosion rate was higher for the AEG MI population as compared to the AEG MX population at the H temperature regime (Chi-square = 242.5, df = 1, P < 0.0001) (Table 1). Lower larval mortality rates among the AEG MI population were measured at higher temperatures (Fisher’s exact test P < 0.0001) (Table 1), while in the AEGMX population, increased temperature resulted in higher mortality rates of larvae (Fisher’s exact test P = 0.02) (Table 1). The mortality rates of larvae were higher among the AEGMX population at the H temperature regime relative to the AEGMI population (Chi-square = 29.81, df = 1, P < 0.0001) (Table 1), however no difference in larval mortality rates were measured between populations at the L temperature regime (Chi-square = 2.320, df = 1, P = 0.1277) (Table 1).
Increase in temperature reduced the pupation rates across both populations [AEGMI (Fisher’s exact test P < 0.0001)] [AEGMX (Fisher’s exact test P < 0.0001)] (Table 1) yet, this effect was larger for the AEGMI population (Chi-square = 38.53, df = 1, P < 0.0001) (Table 1). At the L temperature regime, the AEG MX population had a higher pupation rate compared to the AEG MI population (Chi-square = 22.42, df = 1, P < 0.0001) (Table 1).
A cox proportional hazard model was computed using R, in order to evaluate the impact of temperature, population and sex, as well as the interaction among these variables. Temperature, population and sex all significantly affected eclosion over time. However, there was no interaction between temperature and population (Supplementary file no. 1), (Fig. 1).
Cox proportional hazard model on adult emergence rates across temperature regimes. Ae. aegypti mosquitoes were collected from Miami and Mexico and reared at two different temperature regimen, baseline temperature L (D30N26) and elevated temperature H (D32N28). A cox proportional hazard model to evaluate the effect of population, temperature and sex was designed. Temperature, location and sex are significant factors that affected eclosion over time. However, there was no interaction between temperature and Location as variables. The X axis represents the days post egg eclosion while Y axis represents the probability of eclosion.
The majority of adults reared at the H temperature regime emerged on day 7 (post egg eclosion), as compared to day 8 (post egg eclosion) for the L regime. In all populations, the total number of adults emerging were calculated, and the adult males emerged prior to females. AEGMI H males: 77% emerged on day 7 and 23% on day 8; AEGMI H females: 43% emerged on day 7 and 57% on day 8. AEGMX H males: 77% emerged on day 7 and 23% on day 8; AEGMX H females: 63% emerged on day 7 and 37% on day 8. There were no differences between the populations in the emergence rates (F test P = 0.9205).
It is noteworthy that, more than 50% of the AEGMX L females emerged on the first day when the population recorded its highest adult emergence ratio (77% male and 63% female) (Chi-square = 0.2488, df = 1, P = 0.6179) as compared to the AEGMI L (77% male and 43% female) Chi-square = 16.32, df = 1, P < 0.0001).
Increase in temperature reduces the wing length and adult survivorship
The adults reared at the L temperature regime had a significantly longer wing length than the individuals reared at the H temperature regime [ANOVA, Kruskal–Wallis statistic = 10.72; P (0.0133)] (Fig. 2). A cox proportion hazard model demonstrated that temperature as a variable influenced longevity, however, population did not (Supplementary file 2), (Fig. 3).
Impact of increased temperature on wing length of Ae. aegypti. The impact of increased temperature on the body size was determined by measuring the wing length of the female mosquitoes. Both the Miami and Mexico Ae. aegypti female adults reared at the H temperature regime demonstrated a significantly reduced wing length as compared to the L temperature regime [Kruskal–Wallis statistic = 10.72; P (0.0133)].
The effect of temperature on longevity of Ae. aegypti. The impact of temperature on longevity of the mosquitoes was measured by counting the number of dead adult mosquitoes from the first day of emergence. Increased temperature significantly reduced the longevity of both populations of Ae. aegypti adults (Log-rank [Mantel-Cox] test, Chi-square = 35.66, df = 5, P < 0.0001). A cox proportional hazard model to test the influence of geographical location and temperature as variable on survivability demonstrated that temperature as a variable influenced survivability but location did not influence the model.
The Ae. aegypti populations reared at the H temperature regime had a reduced longevity compared to the L temperature regime (Log-rank [Mantel-Cox] test, Chi-square = 35.66, df = 5, P < 0.0001)29 (Fig. 3). The survival to adulthood was inversely related to temperature [AEGMI (H—41%, L—56%); AEGMX (H—50%, L—78%)].
Skip oviposition behavior is evident across temperature regime and populations
Skip oviposition behavior describes a behavior of Ae. aegypti and Ae. albopictus where they skip from oviposition site to oviposition site to distribute their eggs. The skip oviposition behavior has been observed in some natural populations of Aedes mosquitoes whereby eggs are deposited in more than one oviposition site. Skip oviposition behavior was evident in both temperature regimes of the AEG MI populations.
Increase in temperature did not affect the blood feeding rates of either population [AEGMI (Fisher’s exact test P = 0.0887)], [AEGMX (Fisher’s exact test P = 0.9205)] nor the egg oviposition rates [Kruskal–Wallis test P (0.3916)] (Table 1).
Microbial profiles identified in Aedes midguts
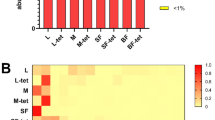
To elucidate the impact of increase in temperature, population and blood meal intake on the midgut microbial profile, we sampled Ae. aegypti from both populations as well as temperature treatments. Total genomic DNA was extracted from adult female Ae. aegypti midguts two days post non-infectious blood meal. As a control, 16S rDNA amplicons were obtained from midguts of adult female fed entirely on 10% sucrose. The sequencing effort generated a total of 6,925,549 reads with a maximum read length of 301 and a mean of 297. After cleaning and trimming the reads, a total of 4,275,936 reads were retained and the frequency of the reads ranged from 25,348–457,857 across the 40 samples analyzed. A total of 3116 OTU were identified. To reduce the potential of alpha and beta diversity biases, the OTU data was rarefied to a sample depth of 34,000 reads per sample. For bacterial profiling, three samples were eliminated from the study as they were below the selected 34,000 reads per sample that all the samples were rarefied to. The OTUs isolated in this study belonged to 5 core phyla: Proteobacteria, Bacteroidetes, Actinobacteria, Cyanobacteria and Firmicutes (Fig. 4).
Relative richness (%) of the core phylotypes identified in Ae. aegypti midguts. The core gut microbiome phyla of Ae. aegypti was Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes and Proteobacteria, respectively. Blood meal acquisition skewed the core phylotypes across the two populations (Bray–Curtis Index, PERMANOVA, F = 2.7182, R2 = 0.072067; P = 0.004).
Temperature significantly impact midgut microbial communities
The midgut microbial abundance and evenness within each population in Miami and Mexico were homogenous in this study (Alpha diversity Shannon diversity Index P = 0.28213; T-test statistic: 1.0924) (Fig. 5B). Increase in temperature did not result in variation in richness or evenness among the individual midguts reared at H and L temperature regimen [Alpha diversity Shannon diversity Index P (0.37668) ANOVA F-value: 1.0663] (Fig. 5C), yet a significant dissimilarity in the midgut microbial community was identified between individuals reared at the H and L temperature regimen (Bray–Curtis Index, PERMANOVA, F = 2.2102; R2 = 0.16731; P = 0.003) (Fig. 6A).
Calculation of the intra-diversity of the gut microbiome of the Aedes mosquitoes. Box plot representation of the diversity of bacterial taxa in Ae. aegypti. (A). Intake of a blood meal resulted in a homogenous microbial midgut both in terms of the richness and evenness [Alpha diversity, Shannon diversity Index P (0.067603; T-test statistic: − 1.8962]. (B) The midgut microbial richness and evenness within each population in Miami and Mexico were homogenous (Alpha diversity Shannon diversity Index P = 0.28213; T-test statistic: 1.0924) and (C) Increase in temperature did not alter the homogeneity in midgut microbiota across the two populations [Alpha diversity Shannon diversity Index P (0.37668) ANOVA F-value: 1.0663]. The X axis represents the combination of the populations and the temperature regimen that they were reared at. The abbreviation of the Aedes aegypti (AEG) population and the temperature regimen that they represent. Such that AEG MI H represents the Ae. aegypti collected from Miami (MI) and reared at the H temperature regimen, AEG MI M- Ae. aegypti collected from Miami (MI) and reared at the L temperature regimen, AEG MX H represents the Ae. aegypti collected from Mexico (MX) and reared at the H temperature regimen while AEG MX L represents the Ae. aegypti collected from Mexico (MX) and reared at the L temperature regimen.
Comparison of bacterial community structure of Ae aegypti mosquitoes. A two-dimensional plot of the Bray–Curtis index. (A) Increase in temperature resulted in significant dissimilarity in the midgut microbial community between the H and L temperature regimen (Bray–Curtis Index, PERMANOVA, F = 2.2063; R2 = 0.16706; P = 0.002). (B) Phylogenetic differences between the two populations (Miami and Mexico) in the bacterial community assemblages was not observed {[PERMANOVA] F- value: 1.4023; R-squared: 0.038523; P (< 0.169)}. (C) Intake of a blood meal altered the microbial community in the midgut of Ae. aegypti individuals (Bray–Curtis Index, PERMANOVA, F = 2.7182, R2 = 0.072067; P = 0.004).
Evenness in phyla distribution of the core microbiota was evident in the AEG MI (L) UNFED individuals [Proteobacteria (0.323), Bacteroidetes (0.158), Actinobacteria (0.075), Cyanobacteria (0.120) and Firmicutes (0.323)] and AEG MX (H) UNFED [Proteobacteria (0.399), Bacteroidetes (0.037), Actinobacteria (0.213), Cyanobacteria (0.130) and Firmicutes (0.222)]. However, AEG MI (H) UNFED [Proteobacteria (0.740) Bacteroidetes (0.191), Actinobacteria (0.060), Cyanobacteria (0.0006) and Firmicutes (0.007)] and AEG MX (L) UNFED [Proteobacteria (0.815), Bacteroidetes (0.022), Actinobacteria (0.037), Cyanobacteria (0.075) and Firmicutes (0.051)] had a skewed core microbiota dominated mainly by Proteobacteria (Fig. 4).
Measures of β-diversity to assess patterns in bacterial community assemblages demonstrated that there are not significant differences between the two populations (Bray–Curtis Index, PERMANOVA, F-value: 1.4001; R-squared: 0.038464; P = 0.169) (Fig. 6B).
Increased temperature resulted in richness of Bacillus subtilis; the richness was notable among the AEG MX_H population (Fig. 7) (Supplementary File 3). Acidovorax citrulli and Pseudomonas aeruginosa were also enriched by increase in temperature and Pseudomonas aeruginosa was most enriched among the AEG MI_H followed by AEG MX_H (Fig. 8) while Dermacoccus was the least enriched of the taxa (Fig. 7). Previous work in our lab has demonstrated that Elizabethkingia thrives better in lower temperature than higher temperatures and this was evident in this study. Stenotrophomonas was highly associated with L regime (Fig. 7).
Enrichment of individual bacterial taxa is associated with a temperature increase in Ae. Aegypti. The Random forest classifier was used to determine the importance of differentially abundant taxa enriched due to increased temperature across both populations and feeding status using the random forest algorithm within Microbiome Analyst. A supervised classification algorithm of trees was created by using bootstrap samples while training data and random feature selection in tree induction. It is an ensemble of unpruned classification or regression trees trained with the bagging method. The Mean Decrease Accuracy obtained by removing the relationship of a taxa and measuring increase in error is reported for each taxon. The taxa with the highest mean decrease in accuracy is considered to have the highest association with the state. Taxa were listed in order of the strength of their level of association with temperature. Stenotrophomonas is highly associated with L regime while Bacillus is strongly associated with H temperature regime. L represents the D30N26 and H represents D32N28 temperature regime. As a taxon approaches red, it indicates richness while the closer it is to blue, the lower its richness in its representation in either H or L temperature regimes.
Blood meal intake did not affect the richness of the midgut microbes but resulted in a heterogenous microbial community in the Ae. aegypti midgut
Blood meal intake significantly altered the homogeneity of the microbial community in the midgut of Ae. aegypti individuals (Bray–Curtis Index, PERMANOVA, F = 2.3422, R2 = 0.36116; P = 0.001) (Fig. 6C). However, the blood meal did not impact the richness and evenness of the midgut microbes [Alpha diversity, Shannon diversity Index P = 0.067603; T-test statistic: − 1.8962] (Fig. 5A). Blood meal ingestion resulted in the gut phyla being dominated mainly by Proteobacteria in both temperature regimes and populations (Fig. 4). The exception was AEG MX (L) gut microbial taxa, which shifted from richness with Proteobacteria (Pseudomonas, Comamonadaceae, Ralstonia and Delftia) (before blood meal ingestion) to Bacteroidetes (Elizabethkingia) (after blood meal) (Fig. 8). Another notable effect of blood meal ingestion was a reduction in the relative richness of Firmicutes (Fig. 4). Taxa for which richness increased significantly following blood feeding have been shown to demonstrate hemolytic activity, including Elizabethkingia30, Pseudomonas31 and Enterobacteriaceae32.
With the exception of AEG MX (L) FED (F) individuals, which demonstrated richness of Elizabethkingia taxa across individuals, variability in dominant taxa at the individual level was identified across groups: (AEG MI (L) F- Elizabethkingia, Chryseobacterium or Enterobacteriaceae; AEG MI (H) F- Ralstonia, Pseudomonas or Elizabethkingia; AEG MX (H) FED – Comamonadaceae, Enterobacteriaceae or Streptococcus) (Fig. 8).
Discussion
This study has demonstrated selective enrichment of heat tolerant taxa (Bacillus) due to temperature increase. Increase in temperature due to climate change is expected to occur in the future33,34. Aedes mosquitoes vector several clinically important viruses, such as Zika virus, chikungunya virus, yellow fever virus and dengue virus35. The microbial community of the mosquito midgut plays a key role in determining the outcome of pathogen infection of mosquitoes22,23,24. Studying the impact of temperature increase on the diversity and function of gut microbial communities is important as it will result in insights on how ectothermic organisms will adapt to climate change in the future36. Studies have shown the activation of heat shock genes due to environmental stress of Bacillus spp37,38. A study with Bacillus spp. SUBB01 at high temperatures estimated that this species could survive at temperatures up to 53 °C39. Further, extracellular fractions of Bacillus spp. could confer protection of heat stressed E. coli. In the present study, Bacillus was the most enriched taxa when Ae. aegypti were reared under increased temperatures. Understanding the mechanism that Bacillus spp. utilize to overcome heat stress will be very important to providing insights into the response of mosquitoes to future climate change.
Several studies have reported the impact of constant temperature increase on the life history traits of mosquitoes40,41,42,43,44,45,46. In nature, daily temperatures vary significantly and this variation likely modulates development rates47. Our study modelled narrower temperature ranges which are within realistic parameters for Ae. aegypti in nature. Our data are consistent with a number of studies demonstrating an inverse relationship between temperature and immature development42,45,47. The effect of temperature on the development and insect morphology has been demonstrated previously in different insects; temperature affects the relationship between wing length and body weight48.
Specifically, the results of our study corroborate a study carried out under field conditions at constant temperature (10–40 °C), for which, survival to adulthood and wing length were inversely related to temperature42. Further, although a study to compare the development time of Ae. aegypti under both constant temperature (12–40 °C) and natural cycling temperature demonstrated that there was no significant difference in development times45, Carrington et al.47 demonstrated that when large fluctuations in temperature are incorporated, development time of Ae. aegypti is accelerated compared to small fluctuations of temperature. Accelerated development equates to additional generations per year and increased opportunity for virus transmission, yet this could to some extent be offset by decreased longevity. The differences between the development time and survival in response to temperature increase suggest a tradeoff, potentially due to failure in enzyme function at high temperatures49.
Consistent with accelerated development, wing length decreased with an increase in temperature in this study. Wing length has been considered a good estimator of body size among Culicids50,51 and a direct relationship between body size and female fecundity exists52,53,54,55.
Better characterization of variations in mosquito fitness in response to different diurnal temperature ranges is needed to better define the role of thermal profiles in determining habitat suitability of this important disease vector.
The results of our study showed a significant dissimilarity in the midgut composition across temperature regimen. Our results are in contrast to several studies that have demonstrated a reduction in the richness of endosymbionts or complete eradication of midgut microbes in insects as a result of temperature increases17,18,19,20. Our studies and those studies demonstrate that temperature has an effect on gut microbes but our results may not translate to other insects sampled in other laboratories or other mosquitoes sampled in this laboratory at a different time. The dissimilarity resulting from temperature increase could significantly impact the vectorial capacity of these populations for the pathogens that they vector.
There were no significant differences in community assemblage across Miami and Mexico populations. However, richness of Elizabethkingia among the Mexican Ae. aegypti relative to the Miami Ae. aegypti was evident. Our previous studies demonstrate that these populations differ in their vector competence, with lower infection and transmission rates for Zika virus particularly at later time points29. Our findings are similar to a study on mosquito microbiome studies in the field across several sites56,57,58,59.
The blood meal did not significantly reduce the richness of the gut microbiota in this study, however it resulted in an altered microbial profile. Our results contrast with other studies reporting a significant alteration of the mosquito midgut microbial richness due to blood meal intake60,61. These differences may be due to the fact that the midgut was obtained 48 h after a blood meal; thus, the midgut microbiome describes very early stages of microbial disruption following a blood meal digestion. Indeed, microbiome have been known to fluctuate in richness throughout the mosquito life cycle62.
Blood meal intake by the mosquitoes resulted in the proliferation of taxa such as Elizabethkingia63,64,65, Comamonadaceae66, Streptococcus66, Ralstonia67 and Chryseobacterium. Our study corroborates the findings of68,69 which demonstrated that E. anophelis is significantly increased after a blood meal uptake, furthermore, E. anophelis facilitates red blood cells lysis with several hemolysins potentially contributing to blood meal digestion in mosquitoes30,70. Understanding the role of these taxa in the vectorial capacity and transmission efficiency of Ae. aegypti could lead to an improved understanding of the pathogen-insect interaction and result in the identification of novel insect control strategies and disease interventions.
Conclusion
This study demonstrated that an increase in temperature resulted in altered life history traits and gut microbial profile of Ae. aegypti. An enrichment of Bacillus, a bacterium that has previously been shown to be associated with heat stress adaptation, was identified. Characterization of the mechanistic relationship of midgut bacteria to vector fitness and vectorial capacity is critical to characterize the impact of future climate change on Ae. aegypti.
Materials and methods
Mosquito rearing
All experimental protocols in this study were approved by Wadsworth Institutional Review Board. This study utilised two populations of Ae. aegypti mosquitoes [Ae. aegypti Miami (AEG MI) and Ae. aegypti Mexico (AEG MX)]. AEG MI, kindly provided by M DeGennaro, Florida International University were collected in Miami-Dade County, Florida in October 2017 and AEG MX provided by GD Ebel’s laboratory at Colorado State University (CSU) were originally collected in Poza Rica, Mexico, in 2016. These two populations were continually maintained at 28 °C with relative humidity of 60% and 16:8 h light: dark (LD) photoperiod at the New York State Department of Health (NYSDOH) Arbovirus Laboratory under standard rearing conditions.
To initiate the life history traits study, the eggs produced by colony females (F23 AEG MX and F3 AEG MI) were hatched at two diurnal temperature regimes: (H) high (day 32 °C/ night 28 °C) and (L) low (day 30 °C/ night 26 °C). Baseline temperature regimes (L) were an estimate of mean day and nighttime temperatures during peak transmission in regions from which populations were derived (www.noaa.gov) while H temperature regime modeled a 2 °C increase in baseline temperature according to the Intergovernmental Panel on Climate Change (IPCC) prediction of 2–4 °C mean global temperature rise over the next century due to global warming71. Temperatures were maintained in Percival environmental chambers (Perry, Iowa). To confirm the temperature and humidity of the chambers, both internal chamber log and independent HOBO data loggers (Onset, Cape Cod, MA) were used. The chambers were maintained at a relative humidity of 70–80%. Before hatching, the water was incubated for 3 h at the two different day time temperatures and the eggs were vacuum hatched in a 1 L conical flask. The vacuum hatching was in order to ensure minimal variation in hatching time. The immature stages of the mosquitoes were reared at the L and H temperature regimes.
One day post hatching, the larvae were immediately transferred to plastic rectangular flat containers [35.6 cm length × 27.9 cm width × 8.3 cm height (Sterilite, catalog no. 1963)] at a density of 200 larvae per 1 L of dechlorinated water and reared at the L and H temperature regimes at 40–60% relative humidity and a LD photoperiod 14:10. This study had three biological replicates which consisted of three different flats in each temperature regimen. Both AEG MX and AEG MI were reared at the H and L temperature regimes. Hence, the life history traits studies were calculated based on the average values. The larvae were fed 0.25 g of Tetra Pond Koi growth feed for 1st and 2nd instar larvae and 0.5 g for 3rd and 4th instar larvae.
Adults were transferred to 3.8 L cardboard cartons, held at the H and L temperature regimes and allowed to mate for 5 days while being provided with sugar and water ad libitum. Water and sugar were withdrawn 24 h before blood feed with a non-infected blood meal.
The mosquitoes were offered a blood meal twice per week, egg cups were placed in the cardboard cartons 48 h after every blood meal in order to collect eggs.
To measure the life history traits, the egg eclosion was recorded daily as well as larvae, pupae and adult survival. Larvae exuviae and cadavers were removed and counted. Larval development was recorded once daily. Due to its biological importance in pathogen transmission, upon the first day of emergence, ten females were randomly picked from each geographical origin and temperature regime. Mosquito wings were measured from the alular notch to the distal margin, excluding the fringe using Axiovision software and Zeiss microscope according to the manufacturer's specifications. The study also compared the blood meal intake across the temperature regimes, the egg clutch size and the hatch rates of the eggs produced.
Blood meal preparation
The non-infectious blood obtained from sheep contained a final concentration of 2.5% sucrose solution was offered to the female mosquitoes after withdrawal of sugar 24 h before blood feed. All methods were carried out in accordance with relevant guidelines and regulations. The females were fed using a 37 °C pre-heated Hemotek membrane feeding system (Discovery Workshops, Acrington, UK) with a porcine sausage casing membrane. After an hour, the mosquitoes were anaesthetized with CO2 and immobilized on a pre-chilled tray connected to 100% CO2. Engorged females were separated and placed in three separate 0.6 L cardboard cartons (30 individuals per carton). Two days after the blood meal, midguts were dissected from female mosquitoes.
DNA isolation and 16S rRNA gene sequencing
Amplification of the 16S rRNA V3-V4 hypervariable region was carried out. PCR reactions were carried out in a total volume of 50 µl, 5 µM of 16S rRNA primers (16S Microbiome_F TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG and 16S Microbiome_R GTC TCG TGG GCT CGG AGA TCT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C), 10 µl genomic DNA and 36 µl AccuStart II PCR supermix (Quanta biosciences, Beverly, USA). The primer sequences include Illumina overhang adapter sequences integrated into them72. The annealing temperature of 55 °C was utilized in this PCR assay. A fragment size of ~ 460 bp of each sample was submitted to Wadsworth Center Applied Genomics Core for sequencing. Automated cluster generation and paired-end sequencing (MiSeq 500 cycle) was performed on the Illumina MiSeq system.
A total of 40 individual midguts were sequenced. Five AEG MX_L fed on 10% sucrose solution (UNFED) and five AEG MX_L fed on a non-infectious blood meal (FED), five AEG MI_L (UNFED), five AEG MI_L (FED), five AEG MX_H (UNFED), five AEG MX_H (FED), five AEG MI_H (UNFED) and five AEG MI_H (FED). A non-template control and a negative control (water) was amplified by PCR (alongside the samples) while the sequencing core spiked the sequencing run with PhiX to check for run quality.
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 5.0. The life history traits, egg hatching rates and blood feeding rates across temperature regimes was calculated using Fisher’s exact test and Chi square test. The study included three biological replicates, hence the statistical analysis of the immature stages was carried out based on the average of the three biological replicates.
Cox proportional hazard model was computed in R, the interactions between variables of geography, temperature and gender and how they affected eclosion and adult survivability were tested, ANOVA test was used to test for the statistical difference.
Data processing and taxonomic assignment
Analysis of the data was carried out on QIITA (https://qiita.ucsd.edu/) and MicrobiomeAnalyst73. In summary, the command split libraries FASTQ was used to convert the de-multiplexed FASTQ files to the format used by QIITA. The samples were trimmed to 100 bp size to allow for meta-analysis. We also set a value for maximum bad run length of 3; maximum barcode errors of 1.5; Phred quality score of 33; minimum per read length fraction of 0.75. A phred quality score above 30 and above bears 0.1% chance of error.
The assignment of the reads to specific taxa was based on a 97% sequence identity to the GreenGenes 16S reference database. A BIOM-formatted OTU table was generated, subsequently. Finally, to visualize the taxonomic profiles of each sample, taxa bar plots were generated using the rarefied data artifact.
Ecological indices and statistical analysis
Ecological indices including richness and diversity (Shannon, Bray–Curtis) were calculated using the MicrobiomeAnalyst73 software. Phylogenetic distance between the gut microbial community was assessed using the unweighted UniFrac distance. Indices were calculated using values from the genus taxonomic rank.
Shannon diversity is utilized to calculate alpha diversity74, Shannon diversity index takes into account both richness and evenness of the microbial species present. It calculates this by utilizing the proportion of species i relative to the total number of species (pi) and multiplying by the natural logarithm of the proportion (lnpi). Shannon Index measures the diversity of the bacterial community within an individual. On the other hand, Bray–Curtis index statistic is used to describe species counts and indicate similarity between individuals rather than difference75. The distance matrix generated after comparison of each sample to the other is visualized for similarity between samples using the Principle Coordinate Analysis ordination method.
To analyze for important taxa species for different experimental factors , random forest algorithm was utilized within Microbiome Analyst73. The default setting of the number of trees to grow and number of predictors to try was applied (500 and 7 respectively) with the randomness setting left on Random forest algorithm76 is a supervised classification algorithm of trees created by using bootstrap samples while training data and random feature selection in tree induction. It is an ensemble of unpruned classification or regression trees trained with the bagging method77.
The accession numbers of the reads sequenced in this study has been deposited in NCBI under the SRA accession numbers: PRNJA605855, PRJNA605852,PRNJA605851, PRNJA605848,PRNJA605849, PRJNA605847, PRNJA605846, PRJNA605844, PRJNA605843, PRJNA605841, PRJNA605840,PRNJA605837, PRJNA605835, PRJNA605834, PRJNA605832, PRJNA605831, PRJNA605830, PRJNA605829, PRJNA605825, PRJNA605824, PRJNA605821, PRNJA605823, PRJNA605819, PRJNA605806, PRJNA605793, PRNJA605788, PRJNA605792, PRJNA605779, PRJNA605785, PRJNA605787, PRJNA605772, PRJNA605773, PRJNA605770, PRJNA605767, PRJNA605764,PRNJA605763, PRJNA605750, PRJNA605761, PRJNA605747.
Data availability
The 16S rRNA reads is available at the NCBI SRA portal https://www.ncbi.nlm.nih.gov/sra.
References
Romero-Lankao, P. et al. Climate change 2014: Impacts, adaptation and vulnerability. In Regional Aspects. Contribution of Working Group II to the Fifth Assesment Report of the Intergovernmental Panel on Climate Change (eds. Barros, V. et al.) 1439–1498 (Cambridge University Press, 2014).
Deutsch, C. et al. Impacts of climate warming on terrestial ectotherms across latitudes. Proc. Natl. Acad. Sci. U. S. A. 105, 6668–6672 (2008).
Tewksbury, J., Huey, R. & Deutsch, C. Ecology-putting the heat on tropical animals. Science (80-) 320, 1296–1297 (2008).
Huey, R. et al. Why tropical forest lizards are vulnerable to climate warming. R. Soc. Lond. Ser. B Biol. Sci. 276, 1939–1948 (2009).
Kearney, M., Shine, R. & Porter, W. The potential for behavioral thermoregulation to buffer ‘cold-blooded’ animals against climate warming. Proc. Natl. Acad. Sci. U. S. A. 106, 3835–3840 (2009).
Thomas, C. et al. Extinction risk from climate change. Nature 427, 145–148 (2004).
Parmesan, C. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Syst. 37, 637–669 (2006).
Etterson, J. & Shaw, R. Constraint to adaptive evolution in response to global warming. Science (80-) 294, 151–154 (2001).
Willis, C., Ruhfel, B., Primack, R., Miller-Rushing, A. & Davis, C. Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc. Natl. Acad. Sci. U. S. A. 105, 170209–217033 (2008).
Davis, C., Willis, C., Primack, R. & Miller-Rushing, A. The importance of phylogeny to the study of phenological response to global climate change. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010, 3201–3213 (2010).
Bhatt, S. et al. The global distribution and burden of dengue. Nature 496, 504–507 (2013).
Wernegreen, J. J. Mutualism meltdown in insects : bacteria constrain thermal adaptation. Curr. Opin. Microbiol. 15, 255–262 (2012).
Breznak, J. A. Intestinal microbiota of termites and other xylophagous insects. Annu. Rev. Microbiol. 36, 323–343 (1982).
Dillon, R. J. & Dillon, V. M. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92 (2004).
Zilber-rosenberg, I. & Rosenberg, E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735 (2008).
Engel, P. & Moran, N. A. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735 (2013).
Hussain, M. et al. Effects of different temperature regimes on survival of Diaphorina citri and its endosymbiotic bacterial communities. Environ. Microbiol. 19, 3439–3449 (2017).
Shan, H. et al. Thermal sensitivity of bacteriocytes constrains the persistence of intracellular bacteria in whitefly symbiosis under heat stress. Environ. Microbiol. 9, 706–716 (2017).
Ross, P. A. et al. Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1006006 (2017).
Prado, S. S., Hung, K. Y., Daugherty, M. P. & Almeida, R. P. P. Indirect effects of temperature on stink bug fitness, via maintenance of gut-associated symbionts. Appl. Environ. Microbiol. 76, 1261–1266 (2010).
Apte-Deshpande, A., Paingankar, M., Gokhale, M. D. & Deobagkar, D. N. Serratia odorifera a midgut inhabitant of aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS ONE 7, e40401 (2012).
Wu, P. et al. A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 25, 101-112.e5 (2019).
Ramirez, J. L. et al. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog. 10, e1004938 (2014).
Ramirez, J. L. et al. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 6, 1–11 (2012).
Abt, M. C. & Pamer, E. G. Commensal bacteria mediated defenses against pathogens. Curr. Opin. Immunol. 29, 16–22 (2014).
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014).
Jupatanakul, N., Sim, S. & Dimopoulos, G. The insect microbiome modulates vector competence for arboviruses. Viruses 6, 4294–4313 (2014).
Caragata, E. P., Tikhe, C. V. & Dimopoulos, G. Curious entanglements: interactions between mosquitoes, their microbiota and arboviruses. Curr. Opin. Virol. 37, 26–36 (2019).
Onyango, M. G. et al. Increased temperatures reduce the vectorial capacity of Aedes mosquitoes for Zika virus. Emerg. Microbes Infect. 9, 67–77 (2020).
Kukutla, P. et al. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector anopheles gambiae. PLoS ONE 9, e97715 (2014).
Vallet-Gely, I. et al. Association of hemolytic activity of pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production †. Appl. Environ. Microbiol. 76, 910–921 (2009).
Gendrin, M. et al. Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nat. Commun. 6, 1–7 (2015).
Epstein, P. R. Is global warming harmful to health?. Sci. Am. 283, 50–57 (2000).
Schellekens, J. et al. A global water resources ensemble of hydrological models: the earthH2Observe Tier-1 dataset. Earth Syst. Sci. Data https://doi.org/10.5194/bgd-9-10669-2012 (2017).
Kraemer, M. U. G. et al. Past and future spread of the arbovirus vectors. Nat. Microbiol. 4, (2019).
Kohl, K. D. & Yahn, J. Effects of environmental temperature on the gut microbial communities of tadpoles. Environ. Microbiol. 18, 1561–1565 (2016).
Schumann, W. The Bacillus subtilis heat shock stimulon. Cell Stress Chaperones 8, 207–217 (2003).
Versteeg, S., Escher, A., Wende, A., Wiegert, T. & Schumann, W. Regulation of the Bacillus subtilis heat shock gene htpG is under positive control. J. Bacteriol. 185, 466–474 (2003).
Munna, S., Tahera, J., Afrad, M. H., Nur, I. T. & Noor, R. Survival of Bacillus spp. SUBB01 at high temperatures and a preliminary assessment of its ability to protect heat—stressed Escherichia coli cells. BMC Res. Notes 8, 1–9 (2015).
Bar-Zeev, M. The effect of temperature on the growth rate and survival of the immature stages of Aëdes aegypti (L.). Bull. Entomol. Res. 49, 157–163 (1958).
Rueda, L., Patel, K., Axtell, R. & Stinner, R. Temperature-dependent development and survival rates of Culex quinquefasciatus and Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 27, 892–898 (1990).
Tun-Lin, W., Burkot, T. & Kay, B. Effects of temperature and larval diet on development rates and survival of the dengue vector Aedes aegypti in north Queensland, Australia. Med. Vet. Entomol. 14, 31–37 (2000).
Costa, E. A. P. de A., Santos, E. M. de M., Correia, J. C. & Albuquerque, C. M. R. de. Impact of small variations in temperature and humidity on the reproductive activity and survival of Aedes aegypti (Diptera, Culicidae). Rev. Bras. Entomol. 54, 488–493 (2010).
Mohammed, A. & Chadee, D. D. Effects of different temperature regimens on the development of Aedes aegypti (L.) (Diptera: Culicidae) mosquitoes. Acta Trop. 119, 38–43 (2011).
Richardson, K., Hoffmann, A. A., Johnson, P., Ritchie, S. & Kearney, M. R. Thermal sensitivity of Aedes aegypti From Australia: empirical data and prediction of effects on distribution. J. Med. Entomol. 48, 914–923 (2011).
Ciota, A., Matacchiero, A., Kilpatrick, A. M. & Kramer, L. D. The effect of temperature on life history traits of culex mosquitoes. J. Med. Entomol. 51, 55–62 (2014).
Carrington, L. B., Armijos, M. V., Lambrechts, L., Barker, C. M. & Scott, T. W. Effects of fluctuating daily temperatures at critical thermal extremes on Aedes aegypti life-history traits. PLoS ONE 8, 1–9 (2013).
Lanciani, C. Photoperiod and the relationship between wing length and body weight in Anopheles quadrimaculatus. J. Am. Mosq. Control Assoc. 8, 297–300 (1992).
Have, T. M. van der. Slaves to the Eyring Equation? Temperature Dependence of Life-History Characters in Developing Ectotherms. (Wageningen University, 2008).
Christophers, S. Aedes aegypti (L.)- the Yellow Fever Mosquito. Its Life History, Bionomics and Structure. (Cambridge University Press, 1960).
Clement, A. The Biology of Mosquitoes. vol 1 (Chapman and Hall, 1992).
Armbruster, P. & Hutchinson, R. A. Pupal mass and wing length as indicators of fecundity in Aedes albopictus and Aedes geniculatus (Diptera: Culicidae). J. Med. Entomol. 39, 699–704 (2002).
Lounibos, L., Nishimura, N., Conn, J. & Lourenco-de-Oliviera, R. Life history correlates of adult size in the malaria vector anopheles darlingi. Mem. Inst. Oswaldo Cruz 90, 769–774 (1995).
Briegel, H. Metabolic relationship between female body size, reserves and fecundity of Aedes aegypti. J. Insect Physiol. 36, 165–172 (1990).
Briegel, H. Fecundity, metabolism, and body size in Anopheles (Diptera: Culicidae), vectors of malaria. J. Med. Entomol. 27, 839–850 (1990).
Osei-Poku, J., Mbogo, C. M., Palmer, W. J. & Jiggins, F. M. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 21, 5138–5150 (2012).
Buck, M. et al. Bacterial associations reveal spatial population dynamics in Anopheles gambiae mosquitoes. Sci. Rep. 6, 1–9 (2016).
Zouache, K. et al. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 75, 377–389 (2010).
Duguma, D., Hall, M. W., Smartt, C. T., Debboun, M. & Neufeld, J. D. Microbiota variations in Culex nigripalpus disease vector mosquito of West Nile virus and Saint Louis Encephalitis from different geographic origins. PeerJ https://doi.org/10.7717/peerj.6168 (2019).
Kumar, S., Molina-Cruz, A., Lalita, G., Janneth, R. & Barillas-Mury, C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science (80-) 327, 1644–1649 (2010).
Terenius, O. et al. Midgut bacterial dynamics in Aedes aegypti. FEMS Microbiol. Ecol. 80, 556–565 (2012).
Villegas, L. E. M. et al. Zika virus infection modulates the bacterial diversity associated with Aedes aegypti as revealed by metagenomic analysis. PLoS ONE 13, e0190352 (2018).
Lau, S. K. P. et al. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg. Infect. Dis. 21, 232–241 (2015).
Lau, S. K. P. et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 6, 8–17 (2016).
Frank, T. et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet 381, 1876 (2013).
Jetté, M. E., Dill-Mcfarland, K. A., Hanshew, A. S., Suen, G. & Thibeault, S. L. The human laryngeal microbiome: effects of cigarette smoke and reflux. Sci. Rep. 6, 6–16 (2016).
Waugh, J., Granger, W. & Gaggar, A. Incidence, relevance and response for ralstonia respiratory infections. Clin. Lab. Sci. 23, 99–106 (2010).
Chen, S., Bagdasarian, M. & Walker, E. D. Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl. Environ. Microbiol. 81, 2233–2243 (2015).
Wang, Y., Gilbreath, T. M., Kukutla, P., Yan, G. & Xu, J. Dynamic gut microbiome across life history of the malaria mosquito anopheles gambiae in Kenya. PLoS ONE 6, 1–9 (2011).
Chen, S., Johnson, B. K., Yu, T., Nelson, B. N. & Walker, E. D. Elizabethkingia anophelis response to iron stress: physiologic, genomic, and transcriptomic analyses. bioRxiv 679894 (2019). https://doi.org/10.1101/679894
IPCC. Third Assessment Report. in (2007).
Zink, S. D., van Slyke, G. A., Palumbo, M. J., Kramer, L. D. & Ciota, A. T. Exposure to west nile virus increases bacterial diversity and immune gene expression in culex pipiens. Viruses 7, 5619–5631 (2015).
Dhariwal, A. et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188 (2017).
Shannon, C. E. A mathematical theory of communication. ACM SIGMOBILE Mob. Comput. Commun. Rev. 5, 3 (1964).
Bray, R. J. & Curtis, J. T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957).
Svetnik, V. et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 43, 1947–1958 (2003).
Liaw, A. & Wiener, M. Classification and regression by randomForest. R news 2, 18–22 (2002).
Acknowledgements
This publication was supported by the Cooperative Agreement Number U01CK000509 funded by the Centres for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centres for Disease Control and Prevention or the Department of Health and Human Services. We would like to extend our gratitude to Dr. Ebel of Colorado State University and Dr. DeGennaro of Florida International University for providing us with Aedes mosquitoes. We would like to thank Dr. Aurélien Vigneron for his kind help with the cox proportional hazard model.
Author information
Authors and Affiliations
Contributions
M.G.O.: Involved in the study design, conducted the experiments, analyzed the results and drafted the paper. B.S.M.: Involved in the mosquito rearing, assisted in mosquito manipulation exercises. P.A.F.: Assisted in mosquito manipulation exercises. M.N.: Involved in the mosquito rearing, assisted in mosquito manipulation exercises. C.A.T.: Involved in the study design, participated in the writing of the paper. K.L.D.: Involved in the study design, participated in the writing of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Onyango, G.M., Bialosuknia, M.S., Payne, F.A. et al. Increase in temperature enriches heat tolerant taxa in Aedes aegypti midguts. Sci Rep 10, 19135 (2020). https://doi.org/10.1038/s41598-020-76188-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76188-x
This article is cited by
-
Effects of heat tolerance on the gut microbiota of Sarcophaga peregrina (Diptera: Sarcophagidae) and impacts on the life history traits
Parasites & Vectors (2023)
-
Vector microbiome: will global climate change affect vector competence and pathogen transmission?
Parasitology Research (2023)
-
Impact of Human Activities on Disease-Spreading Mosquitoes in Urban Areas
Journal of Urban Health (2023)
-
The microbiome and mosquito vectorial capacity: rich potential for discovery and translation
Microbiome (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.