Abstract
Rice bacterial leaf blight is caused by Xanthomonas oryzae pv. oryzae (Xoo) and produces substantial losses in rice yields. Resistance breeding is an effective method for controlling bacterial leaf blight disease. The mutant line H120 derived from the japonica line Lijiangxintuanheigu is resistant to all Chinese Xoo races. To identify and map the Xoo resistance gene(s) of H120, we examined the association between phenotypic and genotypic variations in two F2 populations derived from crosses between H120/CO39 and H120/IR24. The segregation ratios of F2 progeny consisted with the action of a single dominant resistance gene, which we named Xa46(t). Xa46(t) was mapped between the markers RM26981 and RM26984 within an approximately 65.34-kb region on chromosome 11. The 12 genes predicted within the target region included two candidate genes encoding the serine/threonine-protein kinase Doa (Loc_Os11g37540) and Calmodulin-2/3/5 (Loc_Os11g37550). Differential expression of H120 was analyzed by RNA-seq. Four genes in the Xa46(t) target region were differentially expressed after inoculation with Xoo. Mapping and expression data suggest that Loc_Os11g37540 allele is most likely to be Xa46(t). The sequence comparison of Xa23 allele between H120 and CBB23 indicated that the Xa46(t) gene is not identical to Xa23.
Similar content being viewed by others
Introduction
Rice (Oryza sativa) bacterial blight which caused by the pathogen Xanthomonas oryzae pv. oryzae (Xoo) is one of most serious three rice disease in the world, and limits rice productivity each year owing to its high epidemic potential and the lack of effective bactericides1,2. Xoo causes a systemic infection of the vascular system that results in yellowish brown long strip or offwhite lesions along leaf veins at the maximum booting stage. Rice infected by Xoo can lose 10–20% and even up to 80% of its yield3,4. Rice bacterial blight disease is usually prevalent in tropical subtropical regions rice-growing regions except North America5,6.
Normally plant disease resistance is divided into qualitative (complete) or quantitative (partial) according to the plant’s specific interactions against pathogen invasion7. Qualitative resistance belongs to pathogen race-specific resistance which controlled by major resistance (MR) genes. Quantitative resistance belongs to pathogen race-nonspecific resistance which is generally mediated by multiple minor genes or quantitative trait loci (QTLs)8. The rice-Xoo pathosystem as a host–pathogen interactions and co-evolution genetic model was used to dissect plant disease resistance mechanisms5,9. In the rice-Xoo pathosystem, MR-mediated race-specific resistance usually follows the gene-for-gene relationship9,10.
MR has been widely applied to rice breeding in consideration of its high level of resistance and easy genetic manipulation. Application of resistance variety is firmly believed to be the most effective and environment-friendly measure to prevent and control bacterial blight disease1,2. To date, at least 45 race-specific bacterial blight resistance (R) genes to different Xoo races derived from cultivated and wild rice and artificial mutants were identified or mapped11,12. However, resistance provided by R genes could break down due to the emergence of new Xoo races and rapid changes in the pathogenicity of Xoo1,3,10. To solve the problem of Xoo resistance breakdown, new broad-spectrum resistance genes need to be identified.
RNA-seq is used as a standard method for analyzing gene expression profile, including bacterial infections13, such as transcript profiles of the RNA chaperone Hfq in Salmonella enterica14, Burkholderia cenocepacia and Helicobacter pylori15,16 etc. RNA-seq has explicated the difference of RNA level in lots of diverse plants and bacteria caused by diseases17,18. To better understand host plant responses during simultaneous heat and pathogen stress, The experiment that a transcriptomics profile of the Xoo resistance gene Xa7 against Xanthomonas oryzae was conducted during high-temperature stress characterized the plant responses genes against coinstantaneous heat and pathogen stress19. RNA-seq has proved to be obtainable transitorily, yet this method could have already effectively altered our vision of the breadth and depth via eukaryotic transcriptomes assay, which could improve the efficiency of gene identification as well20.
Study on the molecular genetics of mutant lines is an effective approach for new gene discovery and dissection of the biology function mechanism of the plants. In previous research, we identified a new mutant H120 resistant to most of Chinese Xoo races. Purpose of this study was to identify resistance gene in the mutant H120 using the methods of genetic mapping and RNA-seq.
Results
Resistance reaction to Chinese Xoo pathotypes
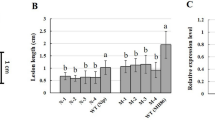
The mutant line H120 which derived from Lijiangxintuanheigu (LTH) (Fig. 1) mutants and the series of varieties including IR24, CO39, LTH, IRBB1, IRBB2, IRBB3, IRBB4, IRBB5, IRBB7, IRBB8, IRBB10, IRBB11, IRBB13, IRBB14, IRBB21 and CBB23 were used for resistance evaluation against six Xoo strains with diverse virulence in South China (Table 1). Among the plants, H120 showed high resistance to all six Xoo pathotypes including pathotype I isolate GD9240, pathotype II GD9269, pathotype III GD9279, pathotype IV GD9315, pathotype V GD9352, and pathotype IX GD9385 from South China. The varieties IR24, CO39, and LTH were all susceptible to all six races. IRBB1, IRBB2, IRBB10, IRBB10 and IRBB14 were resistant to race I and susceptible to other races; IRBB3, IRBB8, IRBB13 and IRBB21 were resistant to race I, II and susceptible to other races; IRBB4 was resistant to race I, II, III, IV and susceptible to other races; IRBB5, IRBB7 and CBB23 were resistant to all the races in this study (Table 1; Fig. 2).
Resistance inheritance of H120
To analyze the resistance inheritance of the mutant line H120, two genetic populations were constructed by crosses among susceptible varieties IR24, CO39 and the resistant parent H120. We used the Xoo predominant race IV isolate GD9315 in South China to inoculate the cross parents and F1 progenies at tillering stage. 20 days after inoculation (DAI), the average lesion lengths of the susceptible parents IR24 and CO39 were 28.2 ± 1.1 cm and 22.2 ± 1.2 cm, respectively, while the average lesion length of H120 was 3.5 ± 0.3 at 20 DAI. F1 plants derived from the IR24/H120 and CO39/H120 crosses all showed resistance to GD9315, whose average lesion length was 3.7 ± 0.3 cm. Mapping populations from the IR24/H120 and CO39/H120 crosses for genetic analysis were used to dissect resistance genetic of H120. We used isolate GD9315 to inoculate 1,263 and 3,128 F2 individuals from the IR24/H120 and CO39/H120 crosses in the field. According to the Standard Evaluation System for Rice, the segregation ratios of resistant and susceptible F2 individuals from the crosses IR24/H120 and CO39/H120 fitted to 3:1 (X2 = 0.0953, P > 0.05 and X2 = 0.3342, P > 0.05, Table 2) with 952 resistant to 311 susceptible and 2,360 resistant to 768 susceptible, respectively, which suggested that H120 harbour a single dominant resistance locus with which temporarily designated Xa46(t).
Molecular identification of the Xa46(t) gene
We used bulked segregant analysis (BSA) and recessive class analysis (RCA) to identify the target gene. The resistant pool (RP) and susceptible pool (SP) with equal DNA of 15 resistant and susceptible F2 individuals were derived from the IR24/H120 cross, respectively. We first used 350 rice simple-sequence repeat (SSR) markers to screen DNA polymorphisms of two parents and the RP and SP. Two SSR markers of chromosome 11, RM26777 and RM206, displayed clear bulk-specific polymorphisms among the resistant and susceptible parents and the RP and SP. These markers were initially used for linkage analysis in the RP and SP individuals, then tested all the F2 individuals. The confirmed results suggested that the Xa46(t) gene was mapped to an 8.0 cM region between RM26777 and RM206.
Preliminarily genetic mapping of Xa46(t)
To map the Xa46(t) gene, we selected a set of 52 SSR markers between markers RM26777 and RM206 to further analyze the linkage with the target gene. Of the 52 SSR markers selected, four SSR markers, RM26801, RM457, RM26834 and RM26988, showed polymorphisms between the IR24 and H120 parents. These four markers were used to analyze the linkage distances to Xa46(t). The confirmed results showed that three polymorphic markers RM26801, RM457, and RM26834 of the RM26777 side were mapped closer to the target gene with 10.0, 8.5 and 6.9 cM. Another marker RM26988 of the RM206 side was mapped more closer to the target gene with 1.0 cM (Fig. 3). Finally the Xa46(t) gene was located in a 7.9 cM zone between RM26834 and RM26988, in which 42 and 6 recombinants were detected from RM26777 and RM206 sites.
Physical mapping of the Xa46(t) gene
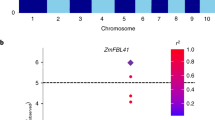
To fine map the Xa46(t) gene, we selected second round of 60 molecular markers to identify linkages to the target gene in the other F2 population from the CO39/H120. A parental polymorphic survey of CO39/H120 was detected first. The test result showed that nine SSR markers with clear polymorphisms were selected as further fine mapping markers within the target region. A total of 702 F2 individuals with highly susceptible reaction (lesion length > 10 cm) to the GD9315 isolate were selected to narrow down the objective region of Xa46(t). Thirty-two BAC/PAC clones from the rice reference genome sequence of Nipponbare were overlapped Xa46(t) within about 3.66 Mb interval (Fig. 4). The anchor markers RM26977, RM26979, RM26981, RM26982, RM26984, and RM26985 were landed on the target PAC clone (P0480H08) at about 98.28 kb physical zone. Recombinants analysis of the 702 F2 individuals showed two and one distinct recombinants were identified with RM26981 and RM26984, in which the target gene was fine mapped to a 65.34-kb on clone P0480H08 between RM26981 and RM26984 (Fig. 4). Within the region of interest. The marker RM26982, flanked by RM26981 and RM26984, co-segregated with Xa46(t). Coincidentally the Xa23(t) gene was also mapped on this region21.
Fine mapping of Xa46(t). aPolymorphisms between H120 (H) and C039 (C) were revealed by SSR markers RM26820, RM26965, RM26977, RM26979, RM26981, RM26982, RM26984, RM26985 and RM26999. Molecular genotypes of some susceptible F2 plants revealed by RM26982 are also shown. bPhysical map of the Xa46(t) locus. Xa46(t) was mapped between the markers RM26981 and RM26984 on chromosome 11 (11S). The nine markers identified in this study, including the co-segregating maker RM26982. Xa46(t) was located in a region corresponding to a 65.34 kb interval in the PAC clone P0480H08 of Nipponbare. Xa23 was also mapped on this region marked with blue (Wang et al. 21. All the linked markers were anchored at the BAC/PAC clones.
Candidate gene annotation of the Xa46(t) gene region
We analyzed the code protein of the Xa46(t) gene mapping rengion based on the release 7.0 of the MSU Rice Genome Annotation Project Database and Resource, which that the target zone contains twelve genes with complete structure (Supplementary Table S1). The genes predicted in the region encode several proteins, including ETO1-like protein 1, the serine/threonine-protein kinase Doa, Calmodulin-2/3/5, ADP-ribosylation factor-like protein 5, an uncharacterized expressed protein, two transposon proteins, and five hypothetical proteins. Of these, we considered the two genes (Loc_Os11g37540 and Loc_Os11g37550) encoding the serine/threonine-protein kinase Doa and Calmodulin-2/3/5 to be promising candidate genes conferring resistance to Xoo isolate GD9315, as that this two coding genes have been linked with resistance response reported22.
Transcriptome analysis of H120 against bacterial blight
We used default parameters on the HISAT software to analyze the filtered sequences for genomic location analysis. The sequencing data quality evaluation showed that RNA-seq was accurate (Supplementary Table S2). The comparison of RNA-seq reads and reference genomes is shown in Supplementary Table S3. Statistics on the density of total mapped reads to each chromosome on the genome (positive and negative chain), explain the relationship between the number of reads on the chromosome and the length of the chromosome. The results showed that the sequence of RNA-seq reads was well-distributed in the rice genome. In RNA-seq analysis, we estimated gene expression level by the count of the sequencing reads of the genomic region or exon of a given gene. The number of genes at different expression levels and the expression level of individual genes were counted (Supplementary Table S4). The FPKM distribution map and violin map were used to compare the gene expression levels under different experimental conditions. For repeated samples under the same experimental conditions, the final FPKM is the average of all repeated data.
Differential expression analysis of candidate genes in Xa46(t) mapping region
The input data for differential gene expression analysis were readcount data from gene expression-level analysis. Within the gene mapping region of the bacterial blight gene Xa46(t), we analyzed the differential expression of genes from the H120 to confirm which candidate is the possible target of Xa46(t). The candidate genes Loc_Os11g37540, Loc_Os11g37560, Loc_Os11g37610 and Loc_Os11g37640 were differentially expressed after inoculation with the pathogen (Fig. 5). Other candidate genes either were not expressed or were expressed at a low level only at a certain timepoint. Combined with the genes’ structure analysis of these four expressed candidate genes, Loc_Os11g37540, the serine/threonine protein kinase Doa, is most likely to be the causal gene for the bacterial blight resistance.
Sequence analysis of Xa23 alleles of H120
To discriminate the relationship of Xa46(t) and Xa23, we sequenced the Xa23 allele of H120, since Xa46(t) was mapped on the Xa23-linked region. First, we amplified the Xa23 allele with several pairs of overlapping gene-specific primers and visualized on agarose gels, the result showed that H120 and CBB23 could amplify the target fragments. Then we sequenced and aligned the PCR products of H120 and analyzed alleles nucleotide diversity polymorphism between H120 and CBB23. We detected 28SNP and 23 InDels through comparative analysis with CLUSTAL W in the total Xa23 gene region (Supplementary Fig. S1). Specially, 3 SNPs and 1 InDel were detected from the Xa23 EBE/ebe domain to the stop codon TAA about 473 bp (Fig. 6). A 6-bp InDel and 1 SNP polymorphism exist in the promoter EBE/ebe region of Xa23 allele between H120 and CBB23, and 1 SNP polymorphism in the CDSXa23 region. Interestingly, the haplotype of Xa23 allele in H120 is almost the same to haplotype H3 of Xa23 allele, in which H3 was defined as susceptible to Xoo23. So the Xa46(t) gene is different from Xa23.
Haplotype analysis of the Xa23 alleles region between H120 and CBB23. The key domain of Xa23/xa23 alleles contains the promoter region − 131 bp upstream sequences from ATG start codon and the 342-bp coding region shown in graphic on the top. The numbers on the top row shows the positions of nucleotide polymorphisms from the EBE/ebe to the stop codon TAA. The “–” indicate deletions.
Discussion
In this study we identified a novel rice Xoo R gene from the mutant line H120. The target gene was mapped to chromosome 11 based on the linkage analysis. Currently about 45 rice bacterial blight resistance genes have been identified or mapped11,12,24,25. Fifteen genes including Xa3/26 , Xa4, Xa10, Xa21, Xa22, Xa23, xa26, Xa30, Xa32, Xa35, Xa36, Xa39, Xa40, Xa43, xa44 and xa44 have been reported to be located on chromosome 1121,24,25,26,27,28,29,30. The target novel gene name Xa46(t) derived from H120, based on the gene nomenclature system of rice Xoo R genes31, located near the centromeric region of chromosome 11.
Of the 45 identified rice bacterial blight resistance genes25, only 11 have been isolated and characterized30,32,33. Four bacterial blight resistance genes, xa5, xa13, xa25 and xa41, are recessive. They possess almost different resistance protein, in which xa5 encodes the small subunit of transcription factor IIA (TFIIAã)34, xa13 and xa25 belong to the MtN3/saliva gene family35 and xa41 represents a new resistance allele owing to an 18-bp deletion in the promoter domain of the OsSWEET14 gene32. Seven dominant bacterial blight resistance genes including Xa1, Xa3/Xa26, Xa4, Xa10, Xa21, Xa23, and Xa27, have been cloned and characterized. Among them, Xa1 encodes an NB-LRR-type protein36, Xa3/Xa26 and Xa21 with LRR receptor kinases (RK)26,37, Xa4 with a cell wall-associated kinase33, Xa10 with a small novel protein harboring 126-amino acid residues and four potential transmembrane helices28, Xa23 with an executor R protein that shares 50% identity to XA1038, and Xa27 with an identical apoplast protein which differ from each other in their promoter regions39. It is very interesting that the functional domains of characterized Xoo R genes vary so widely. In present study, we anlyzed the candidate resistance genes of Xa46(t) between markers RM26981 and RM26984 in the target region based on the information of the MSU Rice Genome Annotation Project Database. Gene prediction results showed that 12 genes were annotated in the linked region. Among them, eight coded for hypothetical proteins, transposon proteins or expressed proteins. No genes encoding transcription factors or proteins similar to known dominant Xoo R gene families mentioned above were identified. We identified two genes, Loc_Os11g37540 and Loc_Os11g37550, encoding the serine/threonine-protein kinase (STPK) Doa and calmodulin-2/3/5 as probable candidate genes for Xa46(t). Some reports indicated that STPKs encoded genes could participate in plant resistance. Such as the barley STPK gene Rpg1 provided resistance to stem rust40, the powdery mildew resistance gene Pm21 encoded STPK protein involved in wheat resistance41 and the rice STPK OsPBL1 potentially involved in rice stripe disease resistance42 etc.. Several studies have shown that the calmodulin-like genes (CMLs) affect plant immune responses. Expression down-regulation of CML NtCaM13 in tobacco could enhanc susceptibility to virulent bacteria Ralstonia solanacearum and fungi Rhizoctonia solani and Pythium aphanidermatum43, whereas overexpression of pepper CMLs CaCaM1 could enhance resistance to the pathogens Xanthomonas campestris pv. Vesicatoria, Pseudomonas syringae and Hyaloperonospora parasitica44. The tomato gene APR134 encoding a CaM-related protein is induced in disease resistance when attacked by Pseudomonas syringae pv. tomato45. Since these reports suggest that STPK and calmodulin-like domain genes might confer resistance against bacterial or fungal pathogens, the two candidates (Loc_Os11g37540 and Loc_Os11g37550) encoding STPK Doa and calmodulin-2/3/5 domain genes were identified as candidates for Xa46(t). Surprisingly, only Loc_Os11g37540 was expressed during challenge with bacterial blight, while Loc_Os11g37550 showed no expression. On the other hand, Xa23(t) is also mapped on this linked region of Nipponbare reference genome, in which LOC_Os11g37620 is regarded as the target candidate resistance gene of Xa23(t)21. Further molecular cloning of Xa23 confirms that it is an executor resistance protein38. Nevertheless the expression data showed that LOC_Os11g37620 was not expressed after inoculation in this study.
The region of chromosome 11 where the Xa46(t) gene mapped to contains at least the Xa23 gene was mapped. To clarify if Xa46(t) is different from Xa23. Sequence comparison of the Xa23 alleles of H120 and CBB23 suggested that the promoter EBE/ebe and CDS regions of Xa23 allele of H120 and LTH are different, in which the EBE/ebe domain of H120 exists 7-bp polymorphism with CBB23, and the CDS domain exists one SNP polymorphism between H120 and CBB23. This important domain information defined H120 as haplotype H3 which was susceptible to Xoo23. So the Xa46(t) gene of H120 is not identical to Xa23, since H120 is a resistant donor. Characterization of Xa46(t) will be helpful to further elucidate the mechanisms of bacterial blight resistance.
Resistance conferred by many bacterial blight and blast R genes can break down when these genes have been widely used for a few years in a large population. Exploitation of more new R genes is urgently needed to solve this problem. The new gene Xa46(t) in this study is expected to be very useful in resistance breeding programs since it is resistant against all the pathotypes in Southern China. Many well-known Xoo R genes, like Xa1, Xa2, Xa10, and Xa14 do not confer resistance to Xoo pathotypes II, III, IV, and V from Southern China; Xa3, xa8, xa13, and Xa21 do not confer resistance to pathotypes III, IV, and V; Xa4 does not confer resistance to pathotype V but does confer resistance to pathotypes I, II, III, and IV. Only xa5 and Xa7 confer strong resistance to pathotypes I, II, III, IV, and V46. As a recessive Xoo resistance gene, xa5 is difficult to use in hybrid breeding. Moreover, Xa4 and Xa7 show specific resistance behaviors related to the receptor background, Xa7 is not suitable for application in hybrid rice, because the F1 generation of its cross with most sterile lines is highly susceptible47. In the current study, the resistance conferred by the dominant target gene Xa46(t) was independent of genetic background. This gene thus has significant value for improving Xoo resistance in hybrid rice, because the F1 generations from crosses with susceptible parents, including sterile lines and inbred rice, remain highly resistant (unpublished data). The Xa46(t) gene in H120 confers resistance to the six Chinese Xoo pathotypes used in this study. We do not know if Xa46(t) confers resistance to non-Chinese Xoo pathotypes because it is difficult to inoculate plants in segregating populations with many pathogen races. Although it is hard to identify Xoo resistant mutants48, we were fortunate to screen out simultaneous plural resistance mutations on one M2 line derived from LTH. Therefore, it is plausible that Xa46(t) confers resistance to all Xoo pathotypes.
Interestingly, Xa gene-mediated resistance was influenced by genetic background. Xa21 was reported that its resistance of transgenic lines enhanced than the gene-donor line37,49. Different rice cultivars carrying Xa3/Xa26 genes showed variable resistance in different indica or japonica genetic backgrounds26. Among the recessive Xoo R genes, xa33(t) and xa42(t) could be influenced by genetic background12. The function of Xa46(t) was not influenced by the background of IR24 and CO39, as their F1 progeny exhibited dominant resistance to all Xoo pathotypes. Thus marker-assisted transfer of the Xa46(t) gene to other genetic backgrounds can be expected to enhance the development of the resistant gene carrier under diverse genetic backgrounds.
Materials and methods
Plant materials
In our previous study, we had conducted a screening on the bacterial blight resistance of different generations of the mutant lines (T0–T8) induced by spaceflight, which is derived from a japonica rice cultivar LTH with highly susceptible to bacterial blight disease. A mutant H120 with highly bacterial blight resistance was obtain, and it showed a stable Xoo resistance from T4 to T8, while its wild type LTH showed highly susceptible to Xoo (Fig. 1). The IR24 and CO39 cultivars were used as susceptible parents in crosses with H120. The F1 progenies from the crosses between H120 and IR24 and CO39 were used to examine whether the Xoo resistance gene acted as a dominant or recessive trait. Two F2 populations were created from the crosses of IR24/H120 and CO39/H120 for fine mapping of the target Xoo resistance gene. The variety LTH, and the International Rice Bacterial Blight (IRBB) near-isogenic lines (NILs) IRBB3, IRBB4, IRBB21, and CBB23 (provided by Chinese Academy of Agricultural Sciences) were used for haplotype analysis. Seeds of these lines were obtained from the International Rice Research Institute, Los Banos, Laguna, Philippines.
Xoo inoculation and evaluation of resistance
Six different virulent strains of Xoo from South China were used for evaluation of resistance: pathotype I isolate GD9240, pathotype II GD9269, pathotype III GD9279, pathotype IV GD9315, pathotype V GD9352, and pathotype IX GD9385. The cultivars H120, IR24, CO39, LTH, IRBB1, IRBB2, IRBB3, IRBB4, IRBB5, IRBB, IRBB8, IRBB10, IRBB11, IRBB13, IRBB21, IRBB14, IRBB21, and CBB23 were inoculated with the Xoo strains by the leaf-clipping method at the maximum tillering stage under field conditions in Guangzhou, South China50. The resistant parent H120, susceptible parents IR24 and CO39, their F1 progenies and the F2 individuals derived from crosses IR24/H120 and CO39/H120 were inoculated with the pathotype IV isolate GD9315 for genetic analysis. The phenotype was evaluated 20 days after inoculation with the Xoo pathogen. The diseased leaf area was evaluated by the Standard Evaluation System for Rice (5th Edition, 2014). In resistance genetic analysis, highly resistant (HR), resistant (R) and moderately resistant (MR) individuals were identified as the resistant phenotype; highly susceptible (HS), susceptible (S) and moderately susceptible (MS) individuals were identified as the susceptible phenotype.
Molecular mapping analysis using simple-sequence repeat markers
DNA from the parents H120, IR24 and CO39 and the two F2 mapping populations of IR24/H120 and CO39/H120 was isolated based on the method of Murray and Thompson51. To map the Xoo resistance gene(s) from H120, 350 simple-sequence repeat (SSR) markers from the Gramene database (https://www.gramene.org; International Rice Genome Sequencing Project 2005) across the 12 rice chromosomes were screened for parental polymorphism. The polymorphism between the donor parent H120 and recipient parents IR24 and CO39 was analyzed following polymerase chain reaction (PCR) with target region markers (Supplementary Table S5). PCR amplification was performed in 20-μl volumes of reaction mixture containing 30–50 ng template DNA, 10 pmol of each primer, and 10 μl of 2X Super Taq PCRMix (Bioteke). The PCR conditions and detected procedure were referred to Chen et al.52.
Markers and target gene linkage analysis
BSA and RCA methods were used to identify polymorphic molecular markers linked to the resistance gene53,54. Information of polymorphic markers listed in Supplementary Table S5. Markers and target gene linkage analysis and map construction were conducted with Mapmaker/Exp (version 3.0) with a threshold LOD score of 3.055. The recombination frequencies and map distances were converted into centiMorgans according to the Kosambi function56.
Analysis of putative candidate genes
The genomic sequence between the flanking SSR markers was downloaded from the reference japonica rice cv. Nipponbare genome released by the International Rice Genome Sequencing Project and analyzed with the software FGENESH (https://www.softberry.com). All genes with clear open reading frames (ORFs) were analyzed on the basis of the available rice genome sequence and annotation databases from NCBI (www.ncbi.nim.nih.org/unigene) and TIGR release 7.1 (https://rice.plantbiology.msu.edu/). Putative functions for these genes defined in the region of interest were annotated using BLAST-P (https://www.ncbi.nlm.nih.gov).
Sample preparation and inoculations for RNA sequencing
Cultures of Xoo isolate GD9315 were grown at 28 °C on peptone sucrose agar (PSA) with 2 µg/ml tetracycline overnight and diluted in sterile distilled water to 108 cfu/ml. Plant leaves were inoculated with dilutions of both strains and water (for mock) using a shearing inoculation method at the 3–4 leaves stage52. Tissue was collected at 0, 6, 12, 24, 48, 72, and 96 h. For bacterial quantification, inoculated leaf tissue was surface sterilized with 10% bleach and rinsed three times with sterile water.
RNA isolation and quantification
Tissue from all of the samples mentioned above was homogenized using mortar and pestle with liquid nitrogen and RNA was purified using the Plant RNA Purification Kit (NucleoZOL, Gene Company). RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Transcriptome sequencing and differential expression analysis
Library preparation and clustering for transcriptome sequencing of all the samples were done by the sequencing company Novogene (China). A total of 1 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample (Novogene Experimental Department). We transferred the original map data to the original sequenced reads with CASAVA base calling through high-throughput sequencing. Phred numerical values were obtained through a probability model in base calling. The clean reads were filtered out from the raw reads for further analysis. The sequencing data quality evaluation is showed in Supplementary Table S1. The index of the reference genome was built using Hisat2 v2.0.4 and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.4. HTSeq v0.9.1 was used to count the read numbers mapped to each gene. FPKM of each gene was calculated based on the length of the gene and read counts mapped to the gene. Differential expression analysis of two conditions was performed using the DEGSeq R package (1.20.0). The P-values were adjusted using the Benjamini & Hochberg method. Corrected P-values of 0.005 and log2 (fold change) of 1 were set as the threshold for significant differential expression. The data analysis of RNA-seq was referred to Conesa et al.57. The analysis was divided into three parts: (1) the readcount was first normalized; (2) then the hypothesis test probability (p value) was calculated according to the model; (3) the last multiple hypothesis test correction was made and we retrieved the FDR value.
Sequence analysis between H120 and CBB23 in the Xa23 gene domain
To distinguish the relationship between the target gene and the Xa23 gene, we sequence the H120’s promoter and exon domain of Xa23 alleles, since the target gene was mapped on the linked Xa23 region. We download the sequence of Xa23 gene including the EBEAvrXa23 (28-bp) and ORF (342-bp) sequences in NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nlh.gov), then designed several pairs of primers (Supplementary Table S6) to amplify a few sections according to the sequence of Xa23 with the software FastPCR 6.5.55. Some of the sequencing confirmed primers derived from the paper of Cui et al.23. CLUSTAL W was used to analyze sequence alignment (https://myhits.sib.swiss/cgi-bin/clustalw) 58. Polymerase Chain Reaction (PCR) was performed in a Gradient iCycler PCR instrument (Bio-Rad) using highly-efficient KOD FX Neo polymerase in a total volume of 25 μl reaction mixture. The reaction mixture contained 50–100 ng of rice genomic DNA (2.0 μl), 10.0 μM of each primer (1.0 μl), 2.0 mM dNTPs (2.0 μl), 2 × PCR KOD buffer (12.5 μl), 0.5 unit (1.0 U/μl) of KOD FX Neo polymerase (0.5 μl), and ddH2O (6.0 μl). The PCR profile consists of 3 min initial denaturation at 94 °C, 35 cycles of amplification with 10 s DNA denaturation at 98 °C, 30 s annealing at 60 °C and a final elongation at 68 °C with 1–2 min depending on the length of different fragment. Subsequently, all amplified products were visualized on 1% agarose gels, and the target fragments were purified with DNA Gel Extraction and Purification Kits and sequenced.
Ethical standards
The experiments comply with the current laws of the country in which they were performed.
References
Khan, A. M., Naeem, M. & Iqbal, M. Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 139, 27–37 (2014).
Mew, T. W., Alvarez, A. M., Leach, J. E. & Swings, J. Focus on bacterial blight of rice. Plant Dis. 77, 5–12 (1993).
Xia, C., Chen, H. & Zhu, H. Identifcation, mapping, isolation of the genes resisting to bacterial blight and breeding application in rice. Mol. Plant Breed. 3, 121–131 (2012).
Srinivasan, B. & Gnanamanickam, S. Identifcation of a new source of resistance in wild rice, Oryza rufpogon, to bacterial blight of rice caused by Indian strains of Xanthomonas oryzae pv. oryzae. Curr. Sci. 88, 25 (2005).
Niño-Liu, D. O., Ronald, P. C. & Bogdanove, A. J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324 (2006).
Ou, S. H. Rice Diseases 2nd edn, 380 (Commonwealth Mycological Institute, Kew, 1985).
Kou, Y. & Wang, S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185 (2010).
Zhang, H. & Wang, S. Rice versus Xanthomonas oryzae pv. oryzae: A unique pathosystem. Curr. Opin. Plant Biol. 16, 188–195 (2013).
Dai, L. Y., Liu, X. L., Xiao, Y. H. & Wang, G. L. Recent advances in cloning and characterization of disease resistance genes in rice. J. Integrat. Plant Biol. 49, 112–119 (2007).
Mew, T. W. Current status and future prospects of research on bacterial blight of rice. Annu. Rev. Phytopathol. 25, 359–382 (1987).
Busungu, C., Taura, S., Sakagami, J. I. & Ichitani, K. Identifcation and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 66, 636–645 (2016).
Liang, L. et al. The rice cultivar Baixiangzhan harbours a recessive gene xa42(t) determining resistance against Xanthomonas oryzae pv. oryzae. Plant Breed. 136, 603–609 (2017).
Westermann, A. J., Gorski, S. A. & Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630 (2012).
Sittka, A. et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4, e1000163 (2008).
Yoder-Himes, D. R. et al. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. USA 106, 3976–3981 (2009).
Sharma, C. M. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010).
Creecy, J. P. & Conway, T. Quantitative bacterial transcriptomics with RNA-seq. Curr. Opin. Microbiol. 23, 33–140 (2015).
Saliba, A. E., Santos, S. C. & Vogel, J. New RNA-seq approaches for the study of bacterial pathogens. Curr. Opin. Microbiol. 35, 78–87 (2017).
Cohen, S. P. et al. RNA-Seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS One 12(11), e0187625 (2017).
Qian, X., Yi, B., Zhuang, Q. F. & Zhong, G. F. RNA-Seq technology and its application in fish transcriptomics. OMICS 18(2), 98–110 (2014).
Wang, C. et al. High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genom. 289, 745–753 (2014).
Reddy, V. S., Safadi, F., Zielinski, R. E. & Reddy, A. S. Interaction of a kinesin-like protein with calmodulin isoforms from Arabidopsis. J. Biol. Chem. 274(44), 31727–33173 (1999).
Cui, H. et al. Promoter variants of Xa23 alleles affect bacterial blight resistance and evolutionary pattern. PLoS One 12(10), e0185925 (2017).
Kim, S. M. Identification of novel recessive gene xa44(t) conferring resistance to bacterial blight races by QTL linkage analysis using an SNP chip. Theoret. Appl. Genet. 131(12), 2733–2743 (2018).
Neelam, K. et al. High-resolution genetic mapping of a novel bacterial glight resistance gene xa-45(t) identified from Oryza glaberrima and transferred to Oryza sativa. Theor Appl Genet. 133(3), 689–705 (2020).
Sun, X. et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzase in rice, encodes an LRR receptor kinase-like protein. Plant J. 37(4), 517–527 (2004).
Xiang, Y., Cao, Y., Xu, C., Li, X. & Wang, S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theoret. Appl. Genet. 113, 1347–1355 (2006).
Tian, D. et al. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 26(1), 497–515 (2014).
Zhang, F. et al. Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 64, 568–575 (2015).
Kim, S. M. et al. Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theoret. Appl. Genet. 128, 1933–1943 (2015).
McCouch, S. R. & Committee on Gene Symbolization, Nomenclature and Linkage, Rice Genetics Cooperative (CGSNL). Gene nomenclature system for rice. Rice 1, 72–84 (2008).
Hutin, M., Sabot, F., Ghesquiere, A., Koebnik, R. & Szurek, B. A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84(4), 694–703 (2015).
Hu, K. et al. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009 (2017).
Iyer, A. S. & McCouch, S. R. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17(12), 1348–1354 (2004).
Liu, Q. et al. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34(11), 1958–1969 (2011).
Yoshimura, S. et al. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 95(4), 1663–1668 (1998).
Song, W. Y. et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806 (1995).
Wang, C. et al. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant. 8(2), 290–302 (2015).
Gu, K. et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435(7045), 1122–1125 (2005).
Brueggeman, R., Drader, T. & Kleinhofs, A. The barley serine/threonine kinase gene Rpg1 providing resistance to stem rust belongs to a gene family with five other members encoding kinase domains. Theoret. Appl. Genet. 113(6), 1147–1158 (2006).
Cao, A. et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 108(19), 7727–7732 (2011).
Lee, K. & Kim, K. The rice serine/threonine protein kinase OsPBL1 (ORYZA SATIVA ARABIDOPSIS PBS1-LIKE 1) is potentially involved in resistance to rice stripe disease. Plant Growth Regul. 77, 67–75 (2015).
Takabatake, R. et al. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 48, 414–423 (2007).
Choi, H. W., Lee, D. H. & Hwang, B. K. The pepper calmodulin gene CaCaM1 is involved in reactive oxygen species and nitric oxide generation required for cell death and the defense response. Mol. Plant Microbe Interact. 22, 1389–1400 (2009).
Chiasson, D., Ekengren, S. K., Martin, G. B., Dobney, S. L. & Snedden, W. A. Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 58, 887–897 (2005).
Zeng, L. et al. Resistance analysis on major resistant genes of rice bacterial blight in different donors and genetic background. Acta Phytopathol. Sin. 46(4), 514–520 (2016) ((in Chinese with English abstract)).
Zeng, L. et al. Resistance of rice near-isogenic lines to bacterial blight strains in South China. Acta Phytopathol. Sin. 36(2), 177–180 (2006) ((in Chinese with English abstract)).
Taura, S., Ogawa, T., Yoshimura, A. & Omura, T. Induction of mutants resistant to bacterial blight in rice. Jpn. J. Breed. 41, 279–288 (1991).
Wang, G. L., Song, W. Y., Ruan, D. L., Sideris, S. & Ronald, P. C. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interact. 9, 850–855 (1996).
Kauffman, H. E., Reddy, A. P. K., Hsieh, S. P. Y. & Merca, S. D. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541 (1973).
Murray, M. G. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326 (1980).
Chen, S. et al. High resolution mapping and gene prediction of Xanthomonas oryzae pv. oryzae resistance gene Xa7. Mol. Breed. 22, 433–441 (2008).
Michelmore, R. N., Paran, I. & Kesseli, R. V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88, 9828–9832 (1991).
Zhang, Q. F. et al. Using bulked extremes and recessive class to mapgenes for photoperiod-sensitive genic male sterility inrice. Proc. Natl. Acad. Sci. USA 91, 8675–8679 (1994).
Lincoln, S., Daly, M. & Lander, E. Constructing genetic maps with Mapmaker/Exp 30 (3rd edn). Whitehead institute technical report, Whitehead Institute, MA (1992).
Kosambi, D. D. The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175 (1944).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016).
Higgins, D. et al. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Acknowledgements
This research is supported by Grants from the Special Fund for Modern Agricultural Industry Technology System of Guangdong Province (2019KJ105), the Talent Training Program Project of Guangdong Academy of Agricultural Sciences, the Earmarked Fund for Modern Agro-Industry Technology Research System (CARS-01-32) and a National key R&D project (2016YFD0300707), the Natural Science Foundation of Guangdong Province (2018B030311035).
Author information
Authors and Affiliations
Contributions
S.C. carried out the development of the mapping population, genetic analysis, primer design, gene mapping, and drafted the manuscript. C.Y.W. participated in the phenotypic selection and data assay of molecular mapping. J.Y.Y. and B.C. performed RNA-seq and candidate genes’ transcription data analysis. W.J.W contributed to the isolation and culture of virulent strains of Xoo used in this study. J.S. and A.Q.F. were involved in phenotypic selection for fine-mapping and data mining of candidates. L.X.Z. and X.Y.Z. designed and coordinated the study, assisted with genetic analyses, and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, S., Wang, C., Yang, J. et al. Identification of the novel bacterial blight resistance gene Xa46(t) by mapping and expression analysis of the rice mutant H120. Sci Rep 10, 12642 (2020). https://doi.org/10.1038/s41598-020-69639-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69639-y
This article is cited by
-
Action mechanism of the potential biocontrol agent Brevibacillus laterosporus SN19-1 against Xanthomonas oryzae pv. oryzae causing rice bacterial leaf blight
Archives of Microbiology (2024)
-
Pyramiding of blast and bacterial blight resistance genes in premium quality rice variety, BRRI dhan63 through marker-assisted breeding approach
Euphytica (2024)
-
Marker-Assisted Simultaneous and Stepwise Pyramiding of Broad-Spectrum Bacterial Leaf Blight Resistance Genes, Xa33 and Xa38, into Salt-Tolerant Rice Variety “CO43”
Plant Molecular Biology Reporter (2024)
-
A review of approaches to control bacterial leaf blight in rice
World Journal of Microbiology and Biotechnology (2022)
-
Marker-assisted introgression of bacterial blight resistance gene xa13 into improved CO43
Euphytica (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.