Abstract
The primary goal of this study was to evaluate the major roles of health-related quality of life (HRQOL) in a 5-year lung cancer survival prediction model using machine learning techniques (MLTs). The predictive performances of the models were compared with data from 809 survivors who underwent lung cancer surgery. Each of the modeling technique was applied to two feature sets: feature set 1 included clinical and sociodemographic variables, and feature set 2 added HRQOL factors to the variables from feature set 1. One of each developed prediction model was trained with the decision tree (DT), logistic regression (LR), bagging, random forest (RF), and adaptive boosting (AdaBoost) methods, and then, the best algorithm for modeling was determined. The models’ performances were compared using fivefold cross-validation. For feature set 1, there were no significant differences in model accuracies (ranging from 0.647 to 0.713). Among the models in feature set 2, the AdaBoost and RF models outperformed the other prognostic models [area under the curve (AUC) = 0.850, 0.898, 0.981, 0.966, and 0.949 for the DT, LR, bagging, RF and AdaBoost models, respectively] in the test set. Overall, 5-year disease-free lung cancer survival prediction models with MLTs that included HRQOL as well as clinical variables improved predictive performance.
Similar content being viewed by others
Introduction
Globally, lung cancer has been the most common cancer for several decades1. Due to advances in early detection and improved treatment strategies1,2, lung cancer mortality has decreased worldwide3, and Korean age-adjusted lung cancer mortality has decreased by 3.4% annually since 20124. With this increase in the number of cancer survivors, it has become important to classify these individuals into precise prognostic groups and provide them with appropriate information for better follow-up planning and personalized self-management.5
Many lung cancer survivors have reported that they had diverse health difficulties2,6, and their health function or symptom burden was more severe than that of others6. In fact, many recent studies have suggested that patient-reported outcomes (PROs), such as health-related quality of life (HRQOL) or clinical data, can provide clear prognostic information7,8. In our previous study of disease-free lung cancer survivors9,10, we found that several HRQOL variables showed prognostic potential, and thus, HRQOL or lifestyle factors can be used to identify patients who could benefit from a specific intervention. Therefore, we aimed to predict lung cancer survivors’ disease-free 5-year survival after primary treatment for lung cancer ended, i.e., the patient survived without any signs or symptoms of that cancer, such as local or regional relapses of the tumor or development of distant metastases, using a combination of sociodemographic, clinical and HRQOL variables.
In general, statistical approaches focus on inferring the characteristics of a population from sample data11, while machine learning techniques (MLTs) may focus on predicting future values by analyzing the given data and have the potential to maximize the prediction accuracy of large clinical data sets12. In addition, MLTs are more suitable for developing prediction models with dozens of parameters when more prognostic variables are included, because standard statistics do not generally work in this situation13. However, although a variety of prediction models based on MLTs for cancer mortality have been developed and utilized in clinical settings14,15, there have been fewer studies regarding the development of MLT-based lung cancer survival prediction models using HRQOL factors.
Here, we proved that the machine learning model including HRQOL data in addition to demographic and clinical parameters was more predictive than existing models that include only demographic and clinical characteristics. We compared the performance of five MLTs by applying each of them to feature set 1 (in which the model considers only demographic and clinical characteristics) and feature set 2 (in which HRQOL factors are added to the variables from feature set 1). The five MLTs used are as follows: decision tree (DT), logistic regression (LR), bagging, random forest (RF), and adaptive boosting (AdaBoost).
Results
Data proportions after data up-sampling and splitting
If the data were well sampled and solved the imbalance problem well, there should be no statistically significant differences in the final comparison of sociodemographic and clinical variables between the deceased and living groups based on the up-sampled data. The final comparison of sociodemographic and clinical variables between the deceased and living groups based on the up-sampled data is shown in Table 1. No statistically significant differences between the deceased and living groups were found after balancing. After missing data imputation and data balancing, the data were split into a training set (80%, n = 1,140) and a validation set (20%, n = 286). There were no significant differences between the training and validation sets.
Importance of the prognostic factors included in the developed prediction model
The importance of the selected prognostic variables was compared with MLT Table 2. The calculated mutual variable importance was normalized, and the sum ranged between 0 and 100%. In feature set 1, cancer stage (II–III) was identified as the most important factor in the DT, bagging, and AdaBoost models. Age was identified as the most important factor in the LR and RF models. In feature set 2, appreciation of life was identified as the most important factor in the DT model, while cancer stage and body mass index (BMI) (kg/m2) before the operation were most important in the bagging model, and sex and anxiety were most important in the RF model. Regional lymph node metastasis and dyspnea were the most important predictors in the AdaBoost model, and personal strength was the most important predictor in the LR model.
Comparisons of the MLT-based models’ performances
Based on the accuracy of the prediction model with cross-validation, each MLT-based prediction model’s performance was measured. The parameters used in each lung cancer survival prediction model are summarized in Table 3, including the validation method with N folds, the training and testing set sizes, the tuning parameter, the performances of the classifiers on the testing set, and the validation results from the fivefold cross-validation dataset using two different feature sets. Among the overall model performances for feature set 1, there were no significant differences in the model accuracies (ranging from 0.647 to 0.713),the LR model had the lowest accuracy, and the RF model had the highest accuracy in feature set 2 (0.746 and 0.916, respectively). Among the models for the fivefold cross-validation sets, the test accuracy of the AdaBoost model exceeded those of the other prognostic models (0.745, 0.825, 0.773, 0.941, and 0.948 for the DT, LR, bagging, RF and AdaBoost models, respectively) in feature set 2.
The receiver operating characteristic (ROC) curves for each feature set of the 5 MLT models based on the cross-validation set that were used to calculate the area under the curve (AUC) were also drawn Fig. 1. Among the models based on feature set 2, the bagging model outperformed the other prognostic models (AUC = 0.850, 0.898, 0.981, 0.966, and 0.949 for the DT, LR, bagging, RF and AdaBoost models, respectively) in fivefold cross-validation. Figure 2 shows the prediction values on the x-axis, and the H-statistic is plotted as five groups in the calibration graphs from the testing set. Four of the models’ calibration plots aligned well with the diagonal lines, although that of the AdaBoost model did not.
Methods
Data acquisition
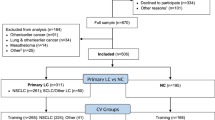
We first identified 2,049 participants aged over 18 years who underwent primary lung cancer surgery between 2001 and 2006 from the Samsung Medical Center or the National Cancer Center in South Korea cancer registries16. The participants were eligible if they (1) were diagnosed with lung cancer (stage 0–III), (2) were treated with curative surgery, and (3) had no evidence of a history of other cancer. We contacted eligible subjects by telephone, and those who agreed to participate were surveyed with the help of our staff at home or in the clinic. In this analysis, we also excluded subjects whose cancer had recurred at that time. As video-assisted thoracic surgery was not often performed from 2001 to 2006, we also excluded patients who received it. Thus, all patients in this study underwent pulmonary resection through open thoracotomy. A total of 1,633 survivors were pathologically diagnosed as disease-free and did not receive any treatment while the study was in progress,906 survivors completed the self-reported survey. After excluding patients with recurring cancer and those for whom the questionnaire was missing, 836 lung cancer survivors were initially included.
Lung cancer patients who did not have evidence of recurrence or death were censored at the last follow-up before the target date. In this study, a regular follow-up was undertaken for each patient based on each hospital’s registry after the completion of treatment. If a patient died during the follow-up, the family caregivers were asked the date of death. Among 836 patients, we excluded 27 subjects whose survival status was censored by December 31, 2011. In total, 809 patients were included in this study. Ethics approval was obtained from the Institutional Review Boards of the National Cancer Center and Samsung Medical Center. The patients eligible to participate were asked to provide informed consent to the staff. Written informed consent was obtained from all the participants before the study. The current study inclusion followed the ethical standards declared in the 1964 Declaration of Helsinki and its later amended version.
Phased feature sets with selected prognostic factors
The study participants’ data included clinical information regarding the primary cancer site, date of cancer diagnosis, cancer stage, treatment type, and other clinical characteristics for all lung cancer survivors. Measuring patients’ symptoms or PROs with a self-reported questionnaire has high validity because asking people directly allows us to reliably obtain their symptom status, and the results can be replicated. Therefore, we collected HRQOL data as well among disease-free lung cancer survivors who were treated with primary lung cancer surgery and survived without cancer recurrence for more than one year through a self-reported questionnaire. Each participant completed the survey including important lung cancer survivorship issues such as HRQOL, anxiety, depression, and posttraumatic growth.
To increase the robustness and validity of a model during the process of prediction modeling, the selection of candidate predictors is important. Final candidate variables that met both the literature review evidence level and statistical significance based on univariate analyses from a previous study16 were selected. (Supplementary Table 1). The variables included demographics (age and sex), socioeconomic status (marital status, educational level, and monthly family income), and past medical history (cancer stage, local invasion of tumor, regional lymph node metastasis, recurrence, comorbidities, treatment type, and time since diagnosis). In addition, lifestyle factors such as BMI and metabolic equivalents of task (MET)-hours per week for physical activity (PA) were considered. For HRQOL assessment, the European Organization for Research and Treatment of Cancer Quality of Life Questionnaire Core 30 (EORTC QLQ-C30)17, the Hospital Anxiety and Depression Scale (HADS)18, and the Posttraumatic Growth Inventory (PTGI) were also selected.
In our model development process, we also calculated a variance inflation factor (VIF) to detect multicollinearity (with a criterion of VIF score greater than 10) in our machine learning models. However, in our data, there were no variables with scores greater than 5, which indicates high correlation in the model; thus, we did not exclude any additional variables from our final candidate variables. In addition, because recurrence was also regarded as an outcome variable, we did not include that variable in the modeling process. For final model construction, we grouped the candidate prognostic factors into two feature sets: (1) sociodemographic and clinical factors and (2) a combination of PROs and lifestyle factors added to the variables from feature set 1.
We defined the endpoint as the fifth year from the date of survey completion after primary treatment for lung cancer ended or the date of any cause of death. To obtain the date of overall survival (OS) after survey completion, we used the National Statistical Office death database linkage through December 31, 2011, as an outcome measure of any cause of death. During the follow-up, we identified 96 deaths (11.9%) and 713 (89.1%) survivals among the 809 subjects. The study design and process is shown in Fig. 3, and the study flow is shown in Supplementary Figure 1.
Statistical analyses
Data preprocessing
Most machine learning algorithms only induce knowledge from the given data, the quality of the extracted knowledge can be determined by the quality of the background data19. Therefore, we first attempted to impute missing values based on important information available in the data set. Although several methodologies are used to treat missing values, we applied the k-nearest neighbor (KNN) algorithm to estimate and substitute our missing data. The KNN algorithm is useful because it can predict both binary and continuous features together. Prior to imputing the missing values, we first investigated the missing numbers of each variable. Then, using the R package “DMwR” and the function “Knn Imputation”, we replaced and imputed the weighted average numbers of the nearest 5 neighbors (k = 5) with missing HRQOL values in our algorithm. The “before” and “after” missing values plotting are shown in the Supplementary Figure 2. The red points are the proportion of missing values, and we could observe that after KNN imputation, there were no more missing values in this data.
Then, to maximize the differences in prognostic strength of the HRQOL scores, we dichotomized each scale of the EORTC QLQ-C30 and EORTC QLQ-LC13 based on the score for the problematic group to investigate clinically meaningful differences: ≤ 33 on a scale of 0–100 for global QOL or functioning scale, and > 66 for symptom scale.16,20 In addition, we used the HADS dichotomized with a cut-off point of 8 (a borderline case of anxiety or depression) as the outcome measure.21 For PTGI, we dichotomized each variable according to the standardized manual22.
In the machine learning classification, important differences in proportions cannot show accurate predictions but also can lead to misleading results. Therefore, in order to make them allowable in real clinical settings, we had to deal with unbalanced problems when the value of finding a few ‘deceased’ classes were much higher than finding a majority. For this preprocessing, we used oversampling to reduce the error costs for the imbalanced data23, and all our study results were based on oversampling. The imbalance in the distribution fails most algorithms from finding a proper solution. The number of ‘deceased’ and ‘living’ cases after oversampling was the equivalent of 713 ‘deceased’ (50%) and ‘living’ (50%) cases. Then, the holdout method was used to randomly split the data sample into two mutually exclusive training (80%) and testing (20%) sets. The training set was utilized to generate the prediction models, and the remaining data were employed as a testing set to estimate the models’ predictive performances.
Machine learning algorithms
Five supervised MLT-based classification models were trained to build each multivariable model to predict the 5-year survival rates for Korean lung cancer long-term survivors in the training set. DT, LR, RF and ensemble learning techniques such as bagging or AdaBoost were selected for the predictive feature selection process. The performances of the derived predictive models based on MLTs were internally validated by fivefold cross-validation.
Individual model learners
The DT and LR models were used for individual model learning. The main components of a DT model are nodes and branches, and the most important steps in building a model are splitting, stopping, and pruning24. In splitting, the purity of the resultant child nodes is used to choose between different potential input variables25,26. A well-classified prediction model shows a higher information gain, and the splitting procedure continues until the stopping criteria are met. Then, we pruned the training set at a point that improved the accuracy of the overall classification and increased the validation error. Each pruning step of the “cp” model can be calculated and plotted as a figure. Finally, DT models from the candidate feature set were developed.
LR is based on a logistic function that estimates the regression equation for a binary (0/1) dependent variable (classification problem). The value of the logit function can be inversely multiplied by the probability of success for the dependent variable so that the survival forecast can be applied to the classification problem. There is a growing perception that simultaneous evaluation of multiple exposures can reduce false-positive findings through several selection methods27. Therefore, we used stepwise selection for the LR model to select the most informative variables. The variable selection was performed in both directions, adding independent variables (forward) and removing previously added variables that were no longer influential (backward). In this stepwise selection process, a sequence of models starting with the null model and ending with the full model was derived. A 5% significance level was chosen as a threshold for the inclusion of a model variable. In this process, we used the generalized linear model (GLM) library from R-3.5.2.
Ensemble feature learners
Ensemble learners such as RF, bagging and adaptive boosting were utilized. Ensemble methods classify data by combining the results of multiple learners to improve classification accuracy by combining predictions from multiple classifiers. Bagging is a technique for generating a large number of training sets by resampling the given learning data with replacement28. With bagging, after generating multiple bootstrap samples, several predictive models are trained for each sample set, and then, the results of each model are combined and predicted. The RF model is a model that adds a random subspace to the bagging29. The difference between a RF model and bagging is that randomly, after choosing ‘m’ variables from among all the variables, the optimal classifier is found by using the ‘m’ selected variables. Finally, the AdaBoost model combines several weak learning algorithms to create a good classification model30. The AdaBoost model sequentially trains the classifiers to complement the weak points of the previous classifier. The R packages caret, randomForest, ipred, and adabag were used for ensemble learning.
Cross-validation
Compared to other models, models built with ensemble techniques are less likely to be overfitted, but it is still something to avoid. Tuning model parameters is one method to prevent overfitting, but it is not the only one. Training features are more likely to lead to overfitting than the model parameters, especially in ensemble learning. Therefore, having a reliable method to check the developed model for overfitting is more important. The choice for the best model based on the k-fold cross-validation results will lead to a model that is not overfitted, which is not necessarily the case for other issues, such as the out-of-bag error. The classifiers of each of the 10 models were trained and evaluated by fivefold cross-validation through the caret package in R.
Model discrimination and calibration
The training and test performance of the MLT algorithms were compared with the model accuracy, which is the proportion of correctly classified samples among the total data. Each predictive performance from the fivefold cross-validation was assessed according to the AUC. The AUCs and 95% confidence intervals (CIs) were calculated to compare the performances of all the proposed models (10 MLT models: 5 from set 1 and 5 from set 2). The normalized variable importance method (VIM)31 was used to determine the importance of explanatory prognostic variables in each of the 10 prediction models. Finally, to estimate the clinical discriminatory capacity of our fivefold cross-validated data sets, we divided the patients into 5 subgroups according to the calculated predictive scores and compared the predictive survival rates to the real-world of survival rates.
Discussion
In this study, we demonstrated the major effects of HRQOL measurements in predicting survival among patients with disease-free lung cancer employing MLT ensemble learners. We also suggested that MLT-based survival prediction models could be used to assist conventional tools in predicting disease-free survivors’ clinical outcomes for lung cancer32 and monitoring their medical status instead of traditional prediction models with individual classification learner-based prediction features16,33. Therefore, we first developed 10 models (5 constructed from only clinical and sociodemographic variables and 5 constructed from clinical or sociodemographic variables and HRQOL data) based on five MLT models (individual learners: DT and LR,ensemble learners: RF, bagging, and AdaBoost) and then compared and validated their prognostic accuracies. Finally, each of the five models based on feature set 2 showed moderately good discrimination and well-calibrated performance compared to the models constructed from only clinical and sociodemographic variables.
Ensemble learning methods showed significantly greater model accuracies (more than 90%) than others in terms of AUC. These machine learning-based lung cancer survival prediction models are the first models developed with not only clinical or sociodemographic factors but also integration of information from multiple factors, such as HRQOL factors combined with clinical factors, ensuring better model performance in terms of both discrimination and calibration and even greater predictive ability than other machine learning models. Among the various MLTs, the RF and AdaBoost models proved superior to the other algorithms.
There are some possible explanations for the findings of this study. First, lung cancer survivors’ HRQOL plays a key role in survival in conjunction with assessments of clinical outcomes, including those based on MLTs. From systematic reviews, we found that there were impressive numbers of studies that showed a positive association between HRQOL and cancer survival. Based on this theoretical background, we constructed a new feature set, which included both clinical variables and HRQOL factors, that quantified good predictive accuracy in our data with five MLTs (DT, LR, bagging, AdaBoost, and RF). From the diverse MLT features, dyspnea, appreciation of life, BMI, anxiety and depression were selected as important variables in addition to cancer stage and sex. Additionally, it is possible that physical function34,35,36,37, dyspnea16,38,39,40,41,42, fatigue34,36,40,43, cough40, anxiety16,41 and depression16,41,42 are strong prognostic factors for survival in lung cancer patients after treatment39,44. Posttraumatic growth factors also have good prognostic value16.
Although biomedical or clinical parameters are generally known as the first factors with prognostic value45, HRQOL parameters have been regarded as additional values in predicting survival.9,34,39 However, even if we cannot change the clinical factors, HRQOL and lifestyle factors can be modified, therefore, we suggested HRQOL parameters as major effects to predict lung cancer survival. Better lung cancer prognostic indices based on both clinical and HRQOL factors need to be developed, and individual assessment algorithms for the prognosis of survivors are essential, guiding the clinical decision-making system to provide more information about their care based on MLTs. These HRQOL findings may indicate disease progression or recurrence that a physical examination by clinicians, a tumor marker evaluation, and imaging studies could not detect.7
In this study, MLTs were identified as having better predictive capabilities in clinical data sets than the traditional approach.12 Additionally, new ensemble learning-based prediction algorithms were more accurate than other MLTs. Although MLTs have been widely used to analyze gene expression data studies46,47 or medical image prediction analyses48, the studies that have explored MLTs in clinical settings are not sufficient, especially with respect to HRQOL. Our approach offers superior performance compared to previous machine learning approaches in predicting cancer survival. In addition, this approach could be used to better stratify lung cancer survivors in future clinical trials of cancer,improving the interpretation of study outcomes or helping identify critical areas could help in the selection of key endpoints for future clinical trials49.
Despite the superior performances of machine learning algorithms, it has been rather limited to use in routine clinical practices because such algorithms cannot be easily calculated with a traditional calculator. DT pruning and LR may produce predictive models with interpretable structures. RF and ensemble learning techniques, such as bagging or AdaBoost, are “black box” models50,51,52, where the function that links the response to the predictive variables is too opaque to use in daily clinical practice. One important advantage of the RF model is that the computational complexity inherent in support-vector machines (SVMs) can be reduced via quadratic optimization. Therefore, for convenience of use in clinical settings, developing a comprehensive digital-based self-management program by including a prediction model can provide more information and help survivors’ decisional support53.
However, our study has several limitations. First, there could be overfitting due to oversampling. It is obvious that the characteristics of the groups are clear, and it is not necessary to perform sampling. Since our data were imbalanced, which affects classification performance, it was necessary to balance the classes by sampling. When a classifier is correctly sampled, the classifier’s performance can be improved through oversampling. If the information in the existing data is lost or distorted during sampling, the learned algorithm does not properly reflect the characteristics of the original data. Therefore, data balancing using SMOTE-NC (non-continuous) to simulate the actual data, including noise that reflects the distribution of the existing data, should be employed in future studies. Second, MLTs that can be adapted to effectively handle survival data should be investigated11. The MLTs that we applied to this study cannot accurately predict the time of an event occurrence, and thus, we could not directly compare them with Cox-based prediction models. Previously, we performed the same process (using original imbalanced data) using Cox proportional hazard regression models, and the prediction model using HRQOL data in addition to clinical and sociodemographic variables was significantly better in terms of C-statistics (Supplementary Table 2). Therefore, including HRQOL with clinical variables together improved predictive performance in both traditional statistical analysis and machine learning techniques as well. Even though, because MLT didn’t consider the time of event, the cox-based model cannot be compared on the same lines. To handle survival problems with MLT, effective algorithms incorporate both statistical methods and MLT, such as survival trees54, random survival forests55, and bagging survival trees28. In addition, the participants were asked at different time intervals relative to the time of their diagnosis. Thus, at the fifth year from the date of survey completion after lung cancer surgery, we adjusted for this time difference by using a covariable that indicates a time since diagnosis. We suggest that an assessment of HRQOL data and lung cancer prediction models based on prognostic factors should be incorporated into routine clinical oncology practice, and further studies, such as randomized controlled trials, should be conducted. Although data from the study suggested that models adding HRQOL data were more accurate, this result should be validated in a wider population. Finally, due to a lack of other cohort sets including PROs among Korean lung cancer survivors, external validation was not conducted. Instead of an external validation set, the entire data set was randomly split to reduce the overfitting in the model then to produce a reliable estimate of the performance of the lung cancer survival model6. Future studies should validate the modeling process with other lung cancer cohort data including PROs.
The current study suggests that socio-clinical variables and HRQOL data can be applied to ensemble MLT algorithms (particularly the RF and AdaBoost algorithms) to predict disease-free lung cancer survival with better predictive performances than models using socioclinical variables only. Most importantly, including both HRQOL and lifestyle factors in a lung cancer survival prediction process with the RF model will provide patients with more accurate information and lower their decisional conflicts. Because cancer survivors need monitoring of multidimensional health-related problems56, the provision of appropriate information through a prediction model is important for better follow-up planning. The improved accuracy of MLT for lung cancer survival prediction can help clinicians and survivors make more clinically meaningful decisions about posttreatment care plans and their support in cancer care.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin 65, 87–108. https://doi.org/10.3322/caac.21262 (2015).
Yun, Y. H. et al. Needs regarding care and factors associated with unmet needs in disease-free survivors of surgically treated lung cancer. Ann. Oncol. 24, 1552–1559. https://doi.org/10.1093/annonc/mdt032 (2013).
Wong, M. C. S., Lao, X. Q., Ho, K. F., Goggins, W. B. & Tse, S. L. A. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci. Rep. 7, 14300. https://doi.org/10.1038/s41598-017-14513-7 (2017).
Jung, K. W., Won, Y. J., Kong, H. J., Lee, E. S. & Community of Population-Based Regional Cancer, R. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2015. Cancer Res. Treat. 50, 303–316. https://doi.org/10.4143/crt.2018.143 (2018).
Simon, R. Clinical trial designs for evaluating the medical utility of prognostic and predictive biomarkers in oncology. Per. Med. 7, 33–47. https://doi.org/10.2217/pme.09.49 (2010).
Yun, Y. H. et al. Health-related quality of life in disease-free survivors of surgically treated lung cancer compared with the general population. Ann. Surg. 255, 1000–1007. https://doi.org/10.1097/SLA.0b013e31824f1e9e (2012).
Gotay, C. C., Kawamoto, C. T., Bottomley, A. & Efficace, F. The prognostic significance of patient-reported outcomes in cancer clinical trials. J. Clin. Oncol. 26, 1355–1363. https://doi.org/10.1200/JCO.2007.13.3439 (2008).
Montazeri, A. Quality of life data as prognostic indicators of survival in cancer patients: an overview of the literature from 1982 to 2008. Health Qual. Life Outcomes 7, 102. https://doi.org/10.1186/1477-7525-7-102 (2009).
Lee, J. Y. et al. Health-Adjusted Life Expectancy (HALE) in Korea: 2005–2011. J. Korean Med. Sci. 31, S139–S145. https://doi.org/10.3346/jkms.2016.31.S2.S139 (2016).
Brown, N. M., Lui, C. W., Robinson, P. C. & Boyle, F. M. Supportive care needs and preferences of lung cancer patients: a semi-structured qualitative interview study. Support. Care in Cancer 23, 1533–1539. https://doi.org/10.1007/s00520-014-2508-5 (2015).
11Wang, P., Li, Y. & Reddy, C. K. Machine learning for survival analysis: a survey. arXiv preprint arXiv:1708.04649 (2017).
Frizzell, J. D. et al. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. JAMA Cardiol. 2, 204–209. https://doi.org/10.1001/jamacardio.2016.3956 (2017).
Cruz, J. A. & Wishart, D. S. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2, 59–77 (2006).
Svensson, C.-M., Hübler, R. & Figge, M. T. Automated classification of circulating tumor cells and the impact of interobsever variability on classifier training and performance. J. Immunol. Res. 2015, 573165 (2015).
Montazeri, M., Montazeri, M., Montazeri, M. & Beigzadeh, A. Machine learning models in breast cancer survival prediction. Technol. Health Care 24, 31–42. https://doi.org/10.3233/THC-151071 (2016).
Yun, Y. H. et al. Prognostic value of quality of life score in disease-free survivors of surgically-treated lung cancer. BMC Cancer 16, 505. https://doi.org/10.1186/s12885-016-2504-x (2016).
Aaronson, N. K. et al. The European Organization for Research and Treatment of Cancer QLQ-C30: a quality-of-life instrument for use in international clinical trials in oncology. J. Natl. Cancer Inst. 85, 365–376 (1993).
Zigmond, A. S. & Snaith, R. P. The hospital anxiety and depression scale. Acta Psychiatr. Scand. 67, 361–370. https://doi.org/10.1111/j.1600-0447.1983.tb09716.x (1983).
Karim, M. N., Reid, C. M., Tran, L., Cochrane, A. & Billah, B. Missing value imputation improves mortality risk prediction following cardiac surgery: an investigation of an Australian patient cohort. Heart Lung Circ. 26, 301–308 (2017).
Fayers, P. M. et al. The EORTC QLQ-C30 Scoring manual 3rd edn. (European Organization for Research and Treatment of Cancer, Brussels, 2001).
Bjelland, I., Dahl, A. A., Haug, T. T. & Neckelmann, D. The validity of the Hospital Anxiety and Depression Scale. An updated literature review. J. Psychosom. Res. 52, 69–77. https://doi.org/10.1016/s0022-3999(01)00296-3 (2002).
Tedeschi, R. G. & Calhoun, L. G. The Posttraumatic Growth Inventory: measuring the positive legacy of trauma. J. Trauma. Stress 9, 455–471. https://doi.org/10.1007/bf02103658 (1996).
Blagus, R. & Lusa, L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinform. 14, 106–106. https://doi.org/10.1186/1471-2105-14-106 (2013).
Wheeler, D. C. et al. Comparison of ordinal and nominal classification trees to predict ordinal expert-based occupational exposure estimates in a case–control study. Ann. Occup. Hyg. 59, 324–335. https://doi.org/10.1093/annhyg/meu098 (2015).
Upadhyay, S. & Patel, N. Study of various decision tree pruning methods with their empirical comparison in WEKA. Int. J. Comput. Appl. 60, 20–25. https://doi.org/10.5120/9744-4304 (2012).
Song, Y. Y. & Lu, Y. Decision tree methods: applications for classification and prediction. Shanghai Arch. Psychiatry 27, 130–135. https://doi.org/10.11919/j.issn.1002-0829.215044 (2015).
Lenters, V., Vermeulen, R. & Portengen, L. Performance of variable selection methods for assessing the health effects of correlated exposures in case-control studies. Occup. Environ. Med. 75, 522–529. https://doi.org/10.1136/oemed-2016-104231 (2018).
Hothorn, T., Lausen, B., Benner, A. & Radespiel-Troger, M. Bagging survival trees. Stat. Med. 23, 77–91. https://doi.org/10.1002/sim.1593 (2004).
Breiman, L. Random forests. Mach. Learn. 45, 5–32. https://doi.org/10.1023/a:1010933404324 (2001).
Schapire, R. E. Empirical inference 37–52 (Springer, Berlin, 2013).
Diaz, I., Hubbard, A., Decker, A. & Cohen, M. Variable importance and prediction methods for longitudinal problems with missing variables. PLoS ONE 10, e0120031. https://doi.org/10.1371/journal.pone.0120031 (2015).
Bouazza, Y. B. et al. Patient-reported outcome measures (PROMs) in the management of lung cancer: a systematic review. Lung Cancer 113, 140–151. https://doi.org/10.1016/j.lungcan.2017.09.011 (2017).
Kumar, S. et al. PrediQt-Cx: post treatment health related quality of life prediction model for cervical cancer patients. PLoS ONE 9, e89851. https://doi.org/10.1371/journal.pone.0089851 (2014).
Fiteni, F. et al. Prognostic value of health-related quality of life for overall survival in elderly non-small-cell lung cancer patients. Eur. J. Cancer 52, 120–128. https://doi.org/10.1016/j.ejca.2015.10.004 (2016).
Maione, P. et al. Pretreatment quality of life and functional status assessment significantly predict survival of elderly patients with advanced non-small-cell lung cancer receiving chemotherapy: a prognostic analysis of the multicenter Italian lung cancer in the elderly study. J. Clin. Oncol. https://doi.org/10.1200/jco.2005.02.527 (2005).
Nowak, A. K., Stockler, M. R. & Byrne, M. J. Assessing quality of life during chemotherapy for pleural mesothelioma: feasibility, validity, and results of using the European Organization for Research and Treatment of Cancer Core Quality of Life Questionnaire and Lung Cancer Module. J. Clin. Oncol. 22, 3172–3180. https://doi.org/10.1200/jco.2004.09.147 (2004).
Langendijk, H. et al. The prognostic impact of quality of life assessed with the EORTC QLQ-C30 in inoperable non-small cell lung carcinoma treated with radiotherapy. Radiother. Oncol. 55, 19–25. https://doi.org/10.1016/s0167-8140(00)00158-4 (2000).
Ban, W. et al. Dyspnea as a prognostic factor in patients with non-small cell lung cancer. Yonsei Med. J. 57, 1063–1069. https://doi.org/10.3349/ymj.2016.57.5.1063 (2016).
Movsas, B. et al. Quality of life supersedes the classic prognosticators for long-term survival in locally advanced non-small-cell lung cancer: an analysis of RTOG 9801. J. Clin. Oncol. 27, 5816–5822. https://doi.org/10.1200/JCO.2009.23.7420 (2009).
Bottomley, A. et al. Symptoms and patient-reported well-being: do they predict survival in malignant pleural mesothelioma? A prognostic factor analysis of EORTC-NCIC 08983: randomized phase III study of cisplatin with or without raltitrexed in patients with malignant pleural mesothelioma. J. Clin. Oncol. 25, 5770–5776. https://doi.org/10.1200/jco.2007.12.5294 (2007).
Nakahara, Y. et al. Mental state as a possible independent prognostic variable for survival in patients with advanced lung carcinoma. Cancer 94, 3006–3015. https://doi.org/10.1002/cncr.10608 (2002).
Wigren, T. Confirmation of a prognostic index for patients with inoperable non-small cell lung cancer. Radiother Oncol 44, 9–15 (1997).
Martins, S. J. et al. Lung cancer symptoms and pulse oximetry in the prognostic assessment of patients with lung cancer. BMC Cancer 5, 72. https://doi.org/10.1186/1471-2407-5-72 (2005).
Sloan, J. A. Metrics to assess quality of life after management of early-stage lung cancer. Cancer J. 17, 63–67. https://doi.org/10.1097/PPO.0b013e31820e15dc (2011).
Paesmans, M. Prognostic and predictive factors for lung cancer. Breathe 9, 112–121. https://doi.org/10.1183/20734735.006911 (2012).
Shin, J. et al. Combined effect of individual and neighborhood socioeconomic status on mortality in patients with newly diagnosed dyslipidemia: a nationwide Korean cohort study from 2002 to 2013. Nutr. Metab. Cardiovasc. Dis. 26, 207–215. https://doi.org/10.1016/j.numecd.2015.12.007 (2016).
Gupta, S. et al. Machine-learning prediction of cancer survival: a retrospective study using electronic administrative records and a cancer registry. BMJ Open 4, e004007. https://doi.org/10.1136/bmjopen-2013-004007 (2014).
Li, C. et al. Using the K-nearest neighbor algorithm for the classification of lymph node metastasis in gastric cancer. Comput. Math. Methods Med. 2012, 876545. https://doi.org/10.1155/2012/876545 (2012).
Mauer, M. et al. The prognostic value of health-related quality-of-life data in predicting survival in glioblastoma cancer patients: results from an international randomised phase III EORTC Brain Tumour and Radiation Oncology Groups, and NCIC Clinical Trials Group study. Br. J. Cancer 97, 302–307. https://doi.org/10.1038/sj.bjc.6603876 (2007).
Burke, H. B. et al. Artificial neural networks improve the accuracy of cancer survival prediction. Cancer 79, 857–862 (1997).
Gao, P. et al. Which is a more accurate predictor in colorectal survival analysis? Nine data mining algorithms vs. the TNM staging system. PLoS ONE 7, e42015 (2012).
Kim, W. et al. Development of novel breast cancer recurrence prediction model using support vector machine. J. Breast Cancer 15, 230–238. https://doi.org/10.4048/jbc.2012.15.2.230 (2012).
Sim, J. A. et al. Perceived needs for the information communication technology (ICT)-based personalized health management program, and its association with information provision, health-related quality of life (HRQOL), and decisional conflict in cancer patients. Psycho-oncology https://doi.org/10.1002/pon.4367 (2017).
Bou-Hamad, I., Larocque, D. & Ben-Ameur, H. A review of survival trees. Stat. Surv. 5, 44–71 (2011).
Ishwaran, H., Kogalur, U. B., Chen, X. & Minn, A. J. Random survival forests for high-dimensional data. Stat. Anal. Data Min. ASA Data Sci. J. 4, 115–132 (2011).
Mewes, J. C., Steuten, L. M. G., Ijzerman, M. J. & van Harten, W. H. Effectiveness of multidimensional cancer survivor rehabilitation and cost-effectiveness of cancer rehabilitation in general: a systematic review. Oncologist 17, 1581–1593. https://doi.org/10.1634/theoncologist.2012-0151 (2012).
Acknowledgements
This work was supported by grants from the National Cancer Center (NCC-0710410 and 1710330) and the NRF (2016H1A2A1907839) of the Republic of Korea. This manuscript was presented as a poster presentation at MED-INFO 2019.
Author information
Authors and Affiliations
Contributions
J.S. and Y.K. participated in study design and coordination, conducted data analyses, participated in the sequence alignment and drafted the manuscript. Y.Y. participated in the design of the study, provided financial support and study materials, collected and assembled the data, interpreted the analyses, participated in the sequence alignment and drafted the manuscript. J.K. participated in the design of the study, participated in the sequence alignment and helped to draft the manuscript. J.L., Y.S., M.K. and J.Z. participated in the design of the study, collected the study participants and helped draft the manuscript. Y.Y. participated in the design of the study, provided financial support and study materials, collected and assembled the data, interpreted the analyses, participated in the sequence alignment and drafted the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sim, Ja., Kim, Y.A., Kim, J.H. et al. The major effects of health-related quality of life on 5-year survival prediction among lung cancer survivors: applications of machine learning. Sci Rep 10, 10693 (2020). https://doi.org/10.1038/s41598-020-67604-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67604-3
This article is cited by
-
Application of the patient-reported outcome-based postoperative symptom management model in lung cancer: a multicenter randomized controlled trial protocol
Trials (2024)
-
Exploring the potential of machine learning in gynecological care: a review
Archives of Gynecology and Obstetrics (2024)
-
Which are best for successful aging prediction? Bagging, boosting, or simple machine learning algorithms?
BioMedical Engineering OnLine (2023)
-
Machine learning (ML) techniques to predict breast cancer in imbalanced datasets: a systematic review
Journal of Cancer Survivorship (2023)
-
Multivariable prediction model of complications derived from diabetes mellitus using machine learning on scarce highly unbalanced data
International Journal of Diabetes in Developing Countries (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.