Abstract
MyoD family inhibitor (MDFI) and MDFI domain-containing (MDFIC) are homologous proteins known to regulate myogenic transcription factors. Hitherto, their role in cancer is unknown. We discovered that MDFI is up- and MDFIC downregulated in colorectal tumors. Mirroring these different expression patterns, MDFI stimulated and MDFIC inhibited growth of HCT116 colorectal cancer cells. Further, MDFI and MDFIC interacted with Jumonji C domain-containing (JMJD) 1 A, a histone demethylase and epigenetic regulator involved in colorectal cancer. JMJD1A influenced transcription of several genes that were also regulated by MDFI or MDFIC. Notably, the HIC1 tumor suppressor gene was stimulated by JMJD1A and MDFIC, but not by MDFI, and HIC1 overexpression phenocopied the growth suppressive effects of MDFIC in HCT116 cells. Similar to colorectal cancer, MDFI was up- and MDFIC downregulated in breast, ovarian and prostate cancer, but both were overexpressed in brain, gastric and pancreatic tumors that implies MDFIC to also promote tumorigenesis in certain tissues. Altogether, our data suggest a tumor modulating function for MDFI and MDFIC in colorectal and other cancers that may involve their interaction with JMJD1A and a MDFIC→HIC1 axis.
Similar content being viewed by others
Introduction
MyoD family inhibitor (MDFI; also known as I-mfa for inhibitor of MyoD family A) was originally cloned as an interaction partner of the MyoD family of myogenic transcription factors. MDFI represses their ability to activate gene transcription likely by two mechanisms: retention of these myogenic factors in the cytoplasm and inhibition of their nuclear DNA binding activity1. Consistently, MDFI is localized in both the cytoplasm and cell nuclei, albeit the predominant localization seems to be within the former1,2. Similar to its inhibitory impact on the MyoD family, MDFI binds to and suppresses the activity of the TCF/LEF-1 transcription factor that is a downstream effector in the WNT/β-catenin signaling pathway. While MDFI inhibits DNA binding of the Xenopus homolog XTcf3, it remains to be studied whether MDFI also diminishes the nuclear function of TCF/LEF-1 through sequestration within the cytoplasm3,4,5. In addition, MDFI binds to β-catenin and this interaction precludes MDFI from binding to MyoD family members, providing a mechanism by which WNT signaling, through increasing β-catenin levels, could overcome the inhibitory effects of MDFI on myogenesis6.
The biological function of MDFI was probed by homozygous deletion of its gene in mice. On a C57Bl/6 background, respective knockout mice die during embryogenesis, which is most likely due to a placental defect. However, on a 129/Sv background, Mdfi−/− mice can survive into adulthood and be fertile; but various degrees of mild spina bifida and skeletal defects affecting the ribs were reported, with the most severe phenotypes causing death shortly after birth7. Another function of MDFI has been observed in osteoclasts: their number is increased and accordingly bone density reduced in Mdfi−/− mice8. Furthermore, suppressing MDFI function through lentivirus-mediated downregulation promoted the regeneration of the murine gastrocnemius muscle after injury, possibly by increasing the activity of the MyoD and myogenin transcription factors9.
A homolog of MDFI is MyoD family inhibitor domain-containing (MDFIC), which also preferentially localizes within the cytoplasm. However, a rare longer MDFIC isoform localizes around and in nucleoli10,11. This isoform may be important to interact with and sequester the HAND1 transcription factor within nucleoli, which is predicted to suppress HAND1-dependent placentation and cardiac morphogenesis12. MDFIC also binds to the glucocorticoid receptor in the cytoplasm, which leads to a change in glucocorticoid receptor phosphorylation. When cells were treated with glucocorticoid, this interaction dissolved and the receptor translocated into the cell nucleus while MDFIC stayed behind in the cytoplasm. Moreover, transcriptome analyses revealed that MDFIC can influence the inflammatory response mediated by the glucocorticoid receptor13. However, no Mdfic-/- mouse model has yet been published that could corroborate these potential functions of MDFIC.
Presently, it is essentially unknown whether MDFI and MDFIC play any role in tumor formation. We found that MDFI and MDFIC are capable of interacting with JMJD1A, a member of the Jumonji C domain-containing (JMJD) protein family. JMJD1A, also called lysine demethylase 3 A (KDM3A), can demethylate di- and monomethylated lysine 9 on histone H314 and may exert pro-oncogenic functions in colon cancer cells15,16,17,18,19. In addition, we discovered changes in the expression pattern of MDFI and MDFIC in colorectal tumors. Hence, we examined the role of MDFI and MDFIC in colorectal cancer cells.
Results
Binding of MDFI and MDFIC to JMJD1A
Pursuing our long-standing interest in the histone demethylase JMJD1A and its interactome20, we also tested if JMJD1A might interact with MDFI or MDFIC. To this end, we performed coimmunoprecipitation experiments. When Flag-tagged MDFI was coexpressed with HA-tagged JMJD1A, MDFI coprecipitated with JMJD1A, but not with the homologous JMJD1B or two other JMJD proteins, UTX and PHF2 (Fig. 1a and Supplementary Fig. S1a). This complex formation between MDFI and JMJD1A was also observable in a reverse order coimmunoprecipitation experiment (Fig. 1b and Supplementary Fig. S1b). Likewise, complex formation was noted between MDFIC and JMJD1A (Fig. 1c,d and Supplementary Fig. S1c,d). Furthermore, when comparable amounts of MDFI and MDFIC were expressed, their degree of complex formation with JMJD1A was similar (Supplementary Fig. S2a).
Interaction of JMJD1A with MDFI and MDFIC. (a) Flag-tagged MDFI was coexpressed with indicated HA-tagged JMJD proteins (JMJD1A, JMJD1B, UTX or PHF2). After anti-HA immunoprecipitation (IP), coprecipitated MDFI was detected by anti-Flag blotting (top panel). The bottom two panels show input levels of Flag- or HA-tagged proteins. (b) Respective reverse order coimmunoprecipitation experiment: anti-Flag IP followed by anti-HA blotting. (c) Coimmunoprecipitation experiments with Flag-MDFIC and HA-tagged JMJD1A, JMJD1B, UTX or SMCX. (d) Corresponding reverse order coimmunoprecipitation experiment with Flag-MDFIC and HA-JMJD1A. (e) Binding of purified, Flag-6His-JMJD1A to comparable amounts of purified GST, GST-MDFI or GST-MDFIC. (f) Coomassie-stained protein gels revealing the purity of respective recombinant proteins. Full-size blots are presented in Supplementary Figs. S1 and S2b,c.
To determine whether JMJD1A binds directly to MDFI and MDFIC, we purified respective proteins and tested their interaction in vitro. Indeed, purified JMJD1A bound to purified MDFI and MDFIC (Fig. 1e,f and Supplementary Fig. S2b,c). Lastly, we observed that MDFI and MDFIC did not interact with the conserved catalytic center of JMJD1A, possibly explaining why not all JMJD proteins are interaction partners of MDFI/MDFIC, while the highly homologous, cysteine-rich C-termini of MDFI and MDFIC were responsible for binding to JMJD1A (Supplementary Fig. S3). Altogether, we conclude that MDFI and MDFIC can directly bind to the histone demethylase JMJD1A.
Opposite changes of MDFI and MDFIC expression in colorectal cancer
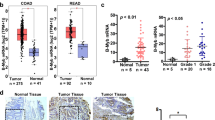
Because JMJD1A is overexpressed in colorectal cancer16,17,18,19 and physically interacts with MDFI/MDFIC, we assessed potential changes of MDFI and MDFIC expression in colorectal tumors by analyzing published microarray data with Oncomine (www.oncomine.org). The Cancer Genome Atlas (TCGA) data set21 revealed that MDFI mRNA was significantly upregulated in cecum, colon, colon mucinous, rectal and rectal mucinous adenocarcinomas compared to normal colon and rectum (Fig. 2a), while MDFIC was on average downregulated (Fig. 2b). Similar MDFI up- and MDFIC downregulation was observed in other microarray data sets, and MDFI appears to be also more expressed in metastatic compared to primary tumor sites (Supplementary Fig. S4). Furthermore, high MDFI mRNA levels were associated with recurrence of colorectal cancer (Fig. 2c; data from reference22), while high MDFIC levels were linked to absence of recurrence (Fig. 2d; data from reference23). Together, these data suggest that MDFI up- and MDFIC downregulation are connected to colorectal tumor formation and possibly also the aggressiveness of the disease.
Altered expression of MDFI and MDFIC in colorectal tumors. (a) MDFI or (b) MDFIC mRNA levels in normal and diseased colorectal tissues. Data were derived from TCGA microarray experiments (reporter A_23_P42165 for MDFI and A_23_P327020 for MDFIC). One-way ANOVA (Dunnett’s multiple comparisons test): *P < 0.05; ***P < 0.001; ****P < 0.0001; ns, not significant. (c) Association of MDFI (reporter 205375_at; data from Lin et al.22) or (d) MDFIC (reporter 1559942_at; data from Jorissen et al.23) mRNA levels with recurrence of colorectal carcinomas; unpaired, two-tailed t test. In all four panels, means with standard deviations are shown.
Impact of MDFI and MDFIC overexpression on HCT116 cells
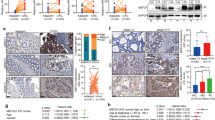
To assess if MDFI or MDFIC can affect cancer cells, we overexpressed these proteins in human HCT116 colorectal cancer cells (Fig. 3a and Supplementary Fig. S5a) and then examined their growth. Of note, MDFI overexpression led to increased cell growth, whereas MDFIC reduced it (Fig. 3b). Moreover, while MDFI overexpression did not robustly affect clonogenic activity, MDFIC strongly suppressed it (Fig. 3c). These results are consistent with the notion that MDFI may promote while MDFIC may inhibit colon cancer formation, which highlights stark differences in function between MDFI and MDFIC despite their homologous amino acid sequences.
Impact of MDFI and MDFIC on cell growth. (a) HCT116 colorectal cancer cells were transduced with retrovirus encoding HA-tagged MDFI or MDFIC. Shown are anti-HA and anti-Lamin B blots. Asterisk marks an unspecific band. Uncropped Western blots are presented in Supplementary Fig. S5a. (b) Growth assays. Two-way ANOVA (Dunnett’s multiple comparison tests); shown are means with standard deviations (n = 3). ****P < 0.0001; ns, not significant. (c) Representative clonogenic assay.
To complement the above overexpression experiments, we also downregulated MDFI or MDFIC with two different shRNAs in HCT116 cells (Supplementary Fig. S6a,d). This did not cause any significant changes in HCT116 cell growth (Supplementary Fig. S6b,e) and also did not affect clonogenic activity (Supplementary Fig. S6c,f). Unfortunately, the unavailability of high-affinity and specific anti-MDFI and anti-MDFIC antibodies did not allow us to measure endogenous MDFI and MDFIC protein levels. If they were very low, any downregulation, although detectable by RT-PCR as shown in Supplementary Fig. S6a,d, would be expected to have no observable impact on HCT116 cells. As such, the results of the shown RNA interference experiments do not allow us to strengthen or refute the hypothesis that MDFI exerts tumor-promoting and MDFIC tumor-suppressing activities.
Transcriptome analysis
To gain more insights into a transcriptional role of MDFI and MDFIC, we created doxycycline-inducible HCT116 cells overexpressing these two proteins. Robust induction of MDFI and MDFIC was observed 12 h after doxycycline addition, and even more so after 36 h (Fig. 4a and Supplementary Fig. S5b). RNA sequencing and cluster analysis of differentially expressed genes revealed that only MDFIC-overexpressing cells were starkly different after 36 h of doxycycline treatment (Fig. 4b and Supplementary Fig. S7). Comparison of RNA sequencing results with RT-PCR data for seven selected genes showed principally consistent trends (Fig. 4c and Supplementary Fig. S5c), thereby validating our RNA sequencing results. This also again emphasized differences between MDFI and MDFIC. For instance, RCAN2 was downregulated upon MDFI overexpression, but not by MDFIC. Or SERPINE1, CTGF, ZEB2 and HIC1 were upregulated by MDFIC, but not by MDFI.
Transcriptome analysis in HCT116 cells. (a) Doxycycline-mediated upregulation of HA-tagged MDFI and MDFIC after 12 and 36 h. Asterisk marks an unspecific band recognized by the anti-HA antibody. Uncropped Western blots are presented in Supplementary Fig. S5b. (b) Hierarchical clustering of 525 differentially expressed genes. (c) Validation of RNA sequencing data (left panel) by RT-PCR (right panel) for indicated target genes after 36 h of doxycycline induction; GAPDH served as a control. Uncropped agarose gels are shown in Supplementary Fig. S5c. (d) Downregulation of JMJD1A with three different shRNAs. Shown are RT-PCR results (top) and Western blots (bottom). Uncropped agarose gels are shown in Supplementary Fig. S9a, and uncropped Western blots in Supplementary Fig. S9b.
We wondered if a differential intracellular localization could be responsible for this different behavior of MDFI and MDFIC. However, MDFI and MDFIC displayed a similar intracellular distribution in HCT116 cells (Supplementary Fig. S8), with both proteins being predominantly in the cytoplasm which is comparable to previously observed data in mouse NIH3T3 fibroblasts or African green monkey COS-1 or COS-7 kidney fibroblast-like cells1,2,10,11. But please note that a small fraction of MDFI and MDFIC was also present in cell nuclei and the insoluble fraction consisting primarily of chromatin, indicating that MDFI and MDFIC are in principle capable of acting as nuclear transcriptional cofactors in HCT116 cells.
We then examined if the above mentioned seven target genes would also be regulated by JMJD1A. To this end, we downregulated JMJD1A with three different shRNAs that led to efficient reduction of JMJD1A protein levels (Fig. 4d, bottom panels, and Supplementary Fig. S9b). With the exception of CTGF, all three JMJD1A shRNAs led to consistent changes in mRNA levels (Fig. 4d, top panels, and Supplementary Fig. S9a), suggesting that PDK4, SERPINE1, TGM2, RCAN2, ZEB2 and HIC1 are JMJD1A target genes. Interestingly, while MDFIC downregulated PDK4, JMJD1A shRNA caused upregulation, and when MDFIC caused upregulation of mRNA levels (SERPINE1, ZEB2, HIC1), JMJD1A depletion had the opposite effect; this suggests that MDFIC and JMJD1A cooperate in the regulation of PDK4, SERPINE1, ZEB2 and HIC1 transcription. Likewise, RCAN2 transcription might be cooperatively repressed by MDFI and JMJD1A. In contrast, JMJD1A downregulation as well as MDFIC overexpression led to TGM2 upregulation, implying antagonistic roles for MDFIC and JMJD1A in TGM2 transcriptional regulation. Overall, these data implicate that JMJD1A can potentially impinge on the transcriptional effects of MDFI and MDFIC.
HIC1 as a potential downstream effector of MDFIC
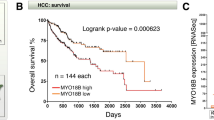
We then focused on one gene strongly activated by MDFIC overexpression, namely HIC1 that encodes for a DNA binding transcriptional repressor. The reasons were that the HIC1 protein is endowed with tumor suppressing activity24 and that HIC1 (which stands for “hypermethylated in cancer 1”) is often epigenetically silenced in colorectal tumors25,26,27,28. Accordingly, we found in published microarray data21,29 that HIC1 mRNA levels are downregulated in colorectal cancer, and low HIC1 levels are associated with reduced survival and increased metastasis (Fig. 5a,b and Supplementary Fig. S10a–c). Of note, consistent with HIC1 transcription being stimulated by MDFIC, MDFIC and HIC1 levels strongly and positively correlated in colon (normal and cancerous) tissue (Fig. 5c and Supplementary Fig. S10d).
HIC1 in colorectal cancer. (a) HIC1 mRNA levels in normal and diseased colorectal tissues. Data were derived from TCGA microarray experiments (reporter A_23_P129856). One-way ANOVA (Dunnett’s multiple comparisons test); ****P < 0.0001. (b) Association of HIC1 mRNA levels (reporter 208461_at; data from Smith et al.29) with survival of colorectal cancer patients; unpaired, two-tailed t test. (c) Correlation between HIC1 and MDFIC mRNA levels across all 234 samples in TCGA data shown in panel a of this figure and panel b of Fig. 2. Pearson correlation: r = 0.5376 (P < 0.0001). (d) Overexpression of HA-tagged HIC1 in HCT116 cells; pQCXIP represents the empty expression vector control. Shown are indicated Western blots. Uncropped Western blots are shown in Supplementary Fig. S9c. (e) Corresponding cell growth assay. Shown are means with standard deviations (n = 3). Two-way ANOVA (Tukey’s multiple comparisons test). ****P < 0.0001; ns, not significant. (f) Representative clonogenic assay.
Next, we overexpressed HIC1 in HCT116 colorectal cancer cells (Fig. 5d and Supplementary Fig. S9c). Reduced cell growth (Fig. 5e) and clonogenic activity (Fig. 5f) were the consequences, mimicking the behavior of overexpressed MDFIC (see Fig. 3). This suggests that upregulation of HIC1 could be an important means for MDFIC to suppress tumorigenesis. Consistently, downregulation of HIC1 blunted the ability of MDFIC to suppress HCT116 cell growth (Supplementary Fig. S11).
MDFI and MDFIC expression in other cancers
Finally, we wondered if MDFI and MDFIC may not only be involved in colorectal cancer, but also in neoplasms of other tissues. Using published microarray data30,31,32, we observed that, identical to colorectal tumors, MDFI and MDFIC exhibited up- and downregulation, respectively, in prostate, breast and ovarian cancer (Fig. 6a,b and Supplementary Fig. S12a). However, both MDFI and MDFIC were upregulated in brain, pancreatic and gastric tumors (Fig. 6c and Supplementary Fig. S12b,c). This implicates that MDFI may generally perform tumor promoting activities, whereas MDFIC might tissue-specifically inhibit or stimulate tumorigenesis. Furthermore, we observed again that HIC1 and MDFIC mRNA levels were strongly correlated in breast, ovarian and gastric cancer and accordingly, like MDFIC, HIC1 was downregulated in breast and ovarian tumors and upregulated in gastric tumors (Supplementary Fig. S13). This reinforces the notion that HIC1 transcription may be regulated by MDFIC.
MDFI and MDFIC mRNA levels in various cancers. (a) MDFI or MDFIC mRNA levels in prostate cancer. Data were derived from Lapointe et al. (reporter IMAGE:33342 for MDFI and IMAGE:148810 for MDFIC)30. Number of specimens is indicated in parentheses. One-way ANOVA (Tukey’s multiple comparisons test). (b) Expression of MDFI and MDFIC in breast tumors; data from Curtis et al. (reporter ILMN_1782798 for MDFI and ILMN_1717366 for MDFIC)31. One-way ANOVA (Dunnett’s multiple comparisons test) was employed to assess differences with normal breast tissue. (c) Analogous in brain tumors; data from Sun et al. (reporter 205375_at for MDFI and 217599_s_at for MDFIC)32. *P < 0.05; ***P < 0.001; ****P < 0.0001; ns, not significant. In all panels, means with standard deviations are shown.
Discussion
In this study, we provide the first evidence that MDFI and MDFIC are involved in colorectal cancer. Perplexingly, despite their high homology, they appear to act in opposite ways, namely MDFI as a tumor promoter and MDFIC as a repressor. This is based on the following evidence: First, we showed that MDFI is overexpressed, while MDFIC is downregulated in colorectal tumors, and high MDFI but low MDFIC levels are associated with more aggressive disease. Second, MDFI was capable of promoting HCT116 colon cancer cell growth, while MDFIC reduced it. Third, MDFI and MDFIC overexpression induced starkly different transcriptome changes. This included upregulation of the HIC1 tumor suppressor gene upon MDFIC overexpression, while MDFI had no impact on HIC1 expression.
Previous studies have shown that MDFI is hypermethylated in colorectal cancer, which would suggest reduced MDFI transcription33,34. This is in contrast to our bioinformatics results, showing upregulation of MDFI in colorectal tumors. However, methylation of only a few CpG sites in the MDFI gene promoter was previously examined, and it is unknown if these CpG sites are crucial for MDFI gene activity. On the other hand, MDFIC has been found to be deleted in myeloid disorders and thereby implied to be a candidate tumor suppressor35, which is in line with our data implicating a tumor suppressing function for MDFIC in the colon.
A second major finding presented in this manuscript is the physical interaction of both MDFI and MDFIC with JMJD1A, a histone demethylase known to perform oncogenic functions in colon cells15,16,17,18,19. Further, JMJD1A and MDFIC (or MDFI) regulated similar genes, and they could act in the same (e.g., PDK4) or opposite (e.g., TGM2) manner with regard to transcriptional activity. This implicates that the interaction of JMJD1A with MDFIC (or MDFI) has different effects on the activity of different promoters. However, this has the caveat that further studies are needed to substantiate that the observed effects of JMJD1A, MDFI or MDFIC on gene transcription were direct and not indirect. In this regard, our and published data1,2,10,11 showed that only a small fraction of MDFI and MDFIC resides in the cell nucleus, while the majority of these proteins is present in the cytoplasm. Hence, MDFI and MDFIC may plausibly perform many functions unrelated to transcriptional coregulation on the chromatin, yet may affect some transcriptional regulators in the cytoplasm. And indeed, previous studies have shown that MDFIC binds to and affects the glucocorticoid receptor in the cytoplasm13, or that MDFI and MDFIC interact with the cytoplasmic AXIN1 protein and in this manner modulate levels of the transcriptional cofactor β-catenin4. It is as well conceivable that MDFI and MDFIC regulate JMJD1A activity in the cytoplasm, since this histone demethylase is also present to a large extent in the cytoplasm of colon cancer cells19 where it may potentially demethylate cytoplasmic, non-histone proteins.
A third finding has been the identification of HIC1 as a seminal downstream effector of MDFIC. Indeed, HIC1 phenocopied MDFIC in suppressing HCT116 cell growth and clonogenic activity, suggesting that a MDFIC→HIC1 axis can restrain colorectal cancer development. Overexpression of MDFIC induced while JMJD1A downregulation decreased HIC1 mRNA levels, suggesting that MDFIC and JMJD1A may cooperate to stimulate HIC1 transcription. However, as neither JMJD1A nor MDFIC are endowed with DNA binding activity, it remains to be determined which HIC1 regulating transcription factor(s) recruits JMJD1A and/or MDFIC to the HIC1 gene locus. Consistent with our data showing that HIC1 overexpression suppressed HCT116 cell growth and clonogenic activity, depletion of Hic1 in mice promoted both polyp formation in cooperation with loss of the tumor suppressor Apc and chemical carcinogenesis in the colon36,37. Mechanistically, HIC1 was shown to repress the SIRT1 gene promoter, thereby preventing SIRT1-mediated deacetylation and inactivation of the tumor suppressor TP5338. However, another way how HIC1 can suppress tumorigenesis independent of TP53 is through averting chromosomal instability39. Therefore, it is tempting to hypothesize that MDFIC also regulates TP53 activity and genome integrity.
Our fourth, puzzling discovery is the fact that MDFIC expression in cancer can be either up- or downregulated depending on the organ. Accordingly, our bioinformatics results imply, but do not prove, that MDFIC context-dependently inhibits (breast, colon, ovary, prostate) or stimulates (brain, pancreas, stomach) tumorigenesis. Interestingly, we noted that MDFI and MDFIC can form homo- as well as heteromers, which is mediated by their conserved cysteine-rich C-terminal domains (Supplementary Fig. S14). Thus, one may speculate that high MDFI levels lead to sequestration of MDFIC into MDFI:MDFIC heteromers and thereby obstruct MDFIC function, while low MDFI levels would allow the formation of MDFIC homomers that only then could inhibit tumorigenesis in the breast, colon, ovary and prostate. Lastly, it is noteworthy that JMJD1A has been implicated in breast, gastric, ovarian and prostate cancer40,41,42,43,44,45, suggesting that the interaction of JMJD1A with MDFI/MDFIC is relevant beyond colorectal tumors.
Methods
Coimmunoprecipitation assay
Expression vectors for indicated proteins were transiently transfected into human 293 T embryonic kidney cells (ATCC CRL-3216) by the calcium phosphate coprecipitation method46. Approximately 40 h later, cells were lysed47 and immunoprecipitations performed as previously described48 with either anti-Flag M2 (Sigma-Aldrich F1804) or anti-HA 12CA5 (Santa Cruz Biotechnology sc-57592) mouse monoclonal antibodies. Immunoprecipitates as well as inputs were boiled in Laemmli sample buffer49 and subjected to polyacrylamide gel electrophoresis50. Separated proteins were transferred to polyvinylidene difluoride membranes51 and challenged with anti-Flag (Sigma-Aldrich F7425) or anti-Myc (Santa Cruz Biotechnology sc-789) rabbit polyclonal antibodies or anti-HA 12CA5 or anti-Myc 9E10 (Sigma-Aldrich M4439) mouse monoclonal antibodies52. This was followed by incubation with horseradish peroxidase-coupled secondary antibodies53 and signal detection through enhanced chemiluminescence54.
In vitro protein binding assay
Fusions of MDFI or MDFIC with glutathione S-transferase (GST) were expressed in Escherichia coli BL21 Codon-Plus (Stratagene)55 and affinity purified with the help of glutathione agarose56. Flag- and 6His-tagged JMJD1A was expressed with the help of baculovirus in Sf9 insect cells (Bac-to-Bac system, Invitrogen) and purified on Ni2+-NTA agarose (Qiagen)57. After binding of GST fusion proteins to glutathione agarose, these beads were challenged with purified JMJD1A58 and bound JMJD1A subsequently revealed by anti-Flag Western blotting59.
Generation of virally transduced cells
Retro-/lentivirus was produced in 293 T cells according to standard procedures60. Then, human HCT116 colorectal cancer cells (ATCC CCL-247) were thrice infected within 24 h61, split 24 h later and selected for two days with 200 µg/ml hygromycin B or 1 µg/ml puromycin62. RNA interference targeted the following sequences within the human JMJD1A open reading frame: GCAGGUGUCAAUAGUGAUA (shRNA #1), GUAGACCUAGUUAAUUGUA (shRNA #2) and CUGCAAAGGACACGGAGAA (shRNA #3). Doxycycline-inducible HCT116 cells were created by viral transduction as described63.
Cell growth and clonogenic assay
2400 virally transduced HCT116 cells were seeded into 96-wells after two days of selection with hygromycin B or puromycin and their growth assessed over the next 1–5 days as described64. Likewise, 2400 virally transduced HCT116 cells were seeded into 6-wells and clonogenic activity revealed approximately 10 days thereafter by staining with crystal violet65.
RNA sequencing and validation
Total RNA was isolated as described19 and then subjected to RNA sequencing at Novogene (https://en.novogene.com). Differential expression analysis of two conditions was performed using version 3.16.5 of the edgeR software package66 and the P values were adjusted using the Benjamini & Hochberg method. Validation of differentially expressed genes was done with RT-PCR67 and visualization of amplified DNA fragments through ethidium-bromide staining68 after agarose gel electrophoresis69. Respective primers are listed in Supplementary Information.
Statistics
Statistical tests that were used are described in the figure legends. Where applicable, means with standard deviations are presented in the figures. All calculations were done with GraphPad Prism 6 for Mac OS X. Statistical significance was assumed for P values less than 0.05.
Data availability
Data, detailed protocols and DNA constructs will be made available upon reasonable request. RNA sequencing data have been deposited in the NCBI BioProject database under accession number PRJNA551463 and can be freely downloaded from the NCBI Sequence Read Archive (accession numbers SRX6406581, SRX6406582, SRX6406583, SRX6406584, SRX6406585, SRX6406586, SRX6406587, SRX6406588 and SRX6406589).
References
Chen, C. M., Kraut, N., Groudine, M. & Weintraub, H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell 86, 731–741 (1996).
Kusano, S., Shiimura, Y. & Eizuru, Y. I-mfa domain proteins specifically interact with SERTA domain proteins and repress their transactivating functions. Biochimie 93, 1555–1564 (2011).
Snider, L. et al. Inhibition of Tcf3 binding by I-mfa domain proteins. Mol. Cell. Biol. 21, 1866–1873 (2001).
Kusano, S. & Raab-Traub, N. I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Mol. Cell. Biol. 22, 6393–6405 (2002).
Pan, W. et al. Beta-catenin relieves I-mfa-mediated suppression of LEF-1 in mammalian cells. J. Cell Sci. 119, 4850–4856 (2006).
Pan, W. et al. Beta-catenin regulates myogenesis by relieving I-mfa-mediated suppression of myogenic regulatory factors in P19 cells. Proc. Natl. Acad. Sci. USA 102, 17378–17383 (2005).
Kraut, N., Snider, L., Chen, C. M., Tapscott, S. J. & Groudine, M. Requirement of the mouse I-mfa gene for placental development and skeletal patterning. EMBO J. 17, 6276–6288 (1998).
Ong, E. C. et al. A TRPC1 protein-dependent pathway regulates osteoclast formation and function. J. Biol. Chem. 288, 22219–22232 (2013).
Hou, L. et al. MiR-27b promotes muscle development by inhibiting MDFI expression. Cell. Physiol. Biochem. 46, 2271–2283 (2018).
Thebault, S., Gachon, F., Lemasson, I., Devaux, C. & Mesnard, J. M. Molecular cloning of a novel human I-mfa domain-containing protein that differently regulates human T-cell leukemia virus type I and HIV-1 expression. J. Biol. Chem. 275, 4848–4857 (2000).
Kusano, S., Yoshimitsu, M., Hachiman, M. & Ikeda, M. I-mfa domain proteins specifically interact with HTLV-1 Tax and repress its transactivating functions. Virology 486, 219–227 (2015).
Martindill, D. M. et al. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat. Cell Biol. 9, 1131–1141 (2007).
Oakley, R. H., Busillo, J. M. & Cidlowski, J. A. Cross-talk between the glucocorticoid receptor and MyoD family inhibitor domain-containing protein provides a new mechanism for generating tissue-specific responses to glucocorticoids. J. Biol. Chem. 292, 5825–5844 (2017).
Yamane, K. et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 (2006).
Krieg, A. J. et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol. Cell. Biol. 30, 344–353 (2010).
Uemura, M. et al. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: in vivo identification from hypoxic tumor cells. Clin. Cancer Res. 16, 4636–4646 (2010).
Li, J. et al. KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/beta-catenin signalling. Nat. Commun. 8, 15146 (2017).
Peng, K. et al. Histone demethylase JMJD1A promotes colorectal cancer growth and metastasis by enhancing Wnt/beta-catenin signaling. J. Biol. Chem. 293, 10606–10619 (2018).
Li, X. et al. A potential common role of the Jumonji C domain-containing 1A histone demethylase and chromatin remodeler ATRX in promoting colon cancer. Oncol. Lett. 16, 6652–6662 (2018).
Knebel, J., De Haro, L. & Janknecht, R. Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J. Cell. Biochem. 99, 319–329 (2006).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Lin, Y. H. et al. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin. Cancer Res. 13, 498–507 (2007).
Jorissen, R. N. et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin. Cancer Res. 15, 7642–7651 (2009).
Wales, M. M. et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat. Med. 1, 570–577 (1995).
Ahuja, N. et al. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res. 57, 3370–3374 (1997).
Pehlivan, S. et al. Gene methylation of SFRP2, P16, DAPK1, HIC1, and MGMT and KRAS mutations in sporadic colorectal cancer. Cancer Genet. Cytogenet. 201, 128–132 (2010).
Abouzeid, H. E. et al. Promoter hypermethylation of RASSF1A, MGMT, and HIC-1 genes in benign and malignant colorectal tumors. Tumour Biol. 32, 845–852 (2011).
Bagci, B. et al. KRAS, BRAF oncogene mutations and tissue specific promoter hypermethylation of tumor suppressor SFRP2, DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients. Cancer Biomark. 17, 133–143 (2016).
Smith, J. J. et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, 958–968 (2010).
Lapointe, J. et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. USA 101, 811–816 (2004).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Sun, L. et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9, 287–300 (2006).
Lin, P. C. et al. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann. Surg. Oncol. 22(Suppl 3), S1419–S1427 (2015).
Li, J. et al. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene 630, 1–7 (2017).
Cigognini, D., Corneo, G., Fermo, E., Zanella, A. & Tripputi, P. HIC gene, a candidate suppressor gene within a minimal region of loss at 7q31.1 in myeloid neoplasms. Leuk. Res. 31, 477–482 (2007).
Mohammad, H. P. et al. Loss of a single Hic1 allele accelerates polyp formation in Apc(Delta716) mice. Oncogene 30, 2659–2669 (2011).
Janeckova, L. et al. HIC1 tumor suppressor loss potentiates TLR2/NF-kappaB signaling and promotes tissue damage-associated tumorigenesis. Mol. Cancer Res. 13, 1139–1148 (2015).
Chen, W. Y. et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123, 437–448 (2005).
Szczepny, A. et al. The tumor suppressor Hic1 maintains chromosomal stability independent of Tp53. Oncogene 37, 1939–1948 (2018).
Wade, M. A. et al. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 43, 196–207 (2015).
Zhao, Q. Y. et al. Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four-stage breast cancer model. Clin. Epigenetics 8, 34 (2016).
Qin, L. et al. The histone demethylase Kdm3a is required for normal epithelial proliferation, ductal elongation and tumor growth in the mouse mammary gland. Oncotarget 8, 84761–84775 (2017).
Yang, H. et al. Elevated JMJD1A is a novel predictor for prognosis and a potential therapeutic target for gastric cancer. Int. J. Clin. Exp. Pathol. 8, 11092–11099 (2015).
Ramadoss, S. et al. Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene 36, 1537–1545 (2017).
Fan, L. et al. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene 35, 2441–2452 (2016).
Goel, A. & Janknecht, R. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol. Cell. Biol. 23, 6243–6254 (2003).
Dowdy, S. C., Mariani, A. & Janknecht, R. HER2/Neu- and TAK1-mediated up-regulation of the transforming growth factor beta inhibitor Smad7 via the ETS protein ER81. J. Biol. Chem. 278, 44377–44384 (2003).
Janknecht, R. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1). Oncogene 22, 746–755 (2003).
Li, X., Moon, G., Shin, S., Zhang, B. & Janknecht, R. Cooperation between ETS variant 2 and Jumonji domain-containing 2 histone demethylases. Mol. Med. Rep. 17, 5518–5527 (2018).
Janknecht, R. & Hunter, T. Activation of the Sap-1a transcription factor by the c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase. J. Biol. Chem. 272, 4219–4224 (1997).
Papoutsopoulou, S. & Janknecht, R. Phosphorylation of ETS transcription factor ER81 in a complex with its coactivators CREB-binding protein and p300. Mol. Cell. Biol. 20, 7300–7310 (2000).
Oh, S., Shin, S., Lightfoot, S. A. & Janknecht, R. 14-3-3 proteins modulate the ETS transcription factor ETV1 in prostate cancer. Cancer Res. 73, 5110–5119 (2013).
Mooney, S. M., Goel, A., D’Assoro, A. B., Salisbury, J. L. & Janknecht, R. Pleiotropic effects of p300-mediated acetylation on p68 and p72 RNA helicase. J. Biol. Chem. 285, 30443–30452 (2010).
Oh, S. & Janknecht, R. Histone demethylase JMJD5 is essential for embryonic development. Biochem. Biophys. Res. Commun. 420, 61–65 (2012).
Wu, J. & Janknecht, R. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J. Biol. Chem. 277, 42669–42679 (2002).
Goel, A. & Janknecht, R. Concerted activation of ETS protein ER81 by p160 coactivators, the acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J. Biol. Chem. 279, 14909–14916 (2004).
Shin, S. et al. Induction of prostatic intraepithelial neoplasia and modulation of androgen receptor by ETS variant 1/ETS-related protein 81. Cancer Res. 69, 8102–8110 (2009).
Kim, T. D. et al. Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J. Clin. Invest. 126, 706–720 (2016).
Kim, J. et al. Histone demethylase JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem. Biophys. Res. Commun. 401, 412–416 (2010).
Kim, T. D. et al. Upregulation of PSMD10 caused by the JMJD2A histone demethylase. Int. J. Clin. Exp. Med. 9, 10123–10134 (2016).
Kim, T. D., Oh, S., Shin, S. & Janknecht, R. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One 7, e34618 (2012).
Kim, T. D. et al. Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon cancer cells and identification of curcuminoids as JMJD2 inhibitors. Am. J. Transl. Res. 6, 236–247 (2014).
Kim, T. D., Shin, S., Berry, W. L., Oh, S. & Janknecht, R. The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J. Cell. Biochem. 113, 1368–1376 (2012).
Kim, T. D., Shin, S. & Janknecht, R. ETS transcription factor ERG cooperates with histone demethylase KDM4A. Oncol. Rep. 35, 3679–3688 (2016).
Berry, W. L., Kim, T. D. & Janknecht, R. Stimulation of beta-catenin and colon cancer cell growth by the KDM4B histone demethylase. Int. J. Oncol. 44, 1341–1348 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Oh, S., Shin, S., Song, H., Grande, J. P. & Janknecht, R. Relationship between ETS transcription factor ETV1 and TGF-beta-regulated SMAD proteins in prostate cancer. Sci. Rep. 9, 8186 (2019).
DiTacchio, L. et al. Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. J. Biol. Chem. 287, 23657–23666 (2012).
Shin, S., Oh, S., An, S. & Janknecht, R. ETS variant 1 regulates matrix metalloproteinase-7 transcription in LNCaP prostate cancer cells. Oncol. Rep. 29, 306–314 (2013).
Acknowledgements
We thank Dr. Leonidas Tsiokas for providing MDFI cDNA and discussions about MDFI function. This work was in part funded by a seed grant from the Stephenson Cancer Center (W.M.F. and R.J.) and through a scholarship from the Graduate School of Jilin University and China-Japan Union Hospital of Jilin University (X.L.). We also thank the Stephenson Cancer Center Molecular Biology Core that has been supported by the National Institutes of Health (grants P20 GM103639 and P30 CA225520). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies, which had no involvement in experimental design and writing this article.
Author information
Authors and Affiliations
Contributions
Y.S., X.L., S.O. and R.J. conceived, designed and performed experiments. Y.S., X.L., S.O., B.Z., W.M.F., S.S. and R.J. analyzed and interpreted data and also contributed to the writing of this manuscript. R.J. supervised this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sui, Y., Li, X., Oh, S. et al. Opposite Roles of the JMJD1A Interaction Partners MDFI and MDFIC in Colorectal Cancer. Sci Rep 10, 8710 (2020). https://doi.org/10.1038/s41598-020-65536-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-65536-6
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.