Abstract
Salmonella enterica ser. Typhimurium monophasic variant 4,[5],12:i:- has been associated with food-borne epidemics worldwide and swine appeared to be the main reservoir in most of the countries of isolation. However, the monomorphic nature of this serovar has, so far, hindered identification of the source due to expansion of clonal lineages in multiple hosts and food producing systems. Since geographically structured genetic signals can shape bacterial populations, identification of biogeographical markers in S. 1,4,[5],12:i:- genomes can contribute to improving source attribution. In this study, the phylogeographical structure of 148 geographically and temporally related Italian S. 1,4,[5],12:i:- has been investigated. The Italian isolates belong to a large population of clonal S. Typhimurium/1,4,[5],12:i:- isolates collected worldwide in two decades showing up to 2.5% of allele differences. Phylogenetic reconstruction revealed that isolates from the same geographical origin form highly supported monophyletic groups, suggesting discrete geographical segregation. These monophyletic groups are characterized by the gene content of a large sopE-containing prophage. Within this prophage, genome-wide comparison identified several genes overrepresented in strains of Italian origin. This suggests that certain lineages may be characterized by the acquisition of specific accessory genetic markers useful for improving identification of the source in ongoing epidemics.
Similar content being viewed by others
Introduction
Salmonella enterica servovar Typhimurium with the antigenic formula 4,[5],12:i:- is considered a monophasic variant of S. Typhimurium (MVSTm) lacking the second phase flagellar antigen1. MVSTm has recently emerged in food-borne epidemics of multi-drug resistance (MDR) strains responsible for several outbreaks in Europe (EU)2 as well as in other continents1. Since this serovar was detected, as far back in 19973, it has been repeatedly associated to humans and swine production, but also to environmental samples and other food-producing animals, such as avian and cattle2,4,5,6,7,8. The increasing spread of MVSTm in EU, the growing number of food-borne outbreaks in recent years6 and the difficulties in identifying the source due to the monomorphic nature of this serovar continue to be a public health concerns. The existence of at least two distinct clones (European and Spanish clone) emerging independently from ancestral S. Typhimurium strains has been previously described2,5,9,10,11. Additionally, different antimicrobial resistance (AR) patterns have been associated to both clones. The prevalence of simultaneous resistance to ampicillin, streptomycin/spectinomycin, sulphonamides and tetracycline (R-type ASSuT) has been described in strains from EU clone12, while in strains from the Spanish clone an additional resistance to chloramphenicol, gentamycin and trimethoprim has been reported13.
It has been argued that traditional typing methods are not well suited to unravel the evolution dynamics of MVSTm population, as well as the source attribution and epidemiology of this monomorphic bacterial pathogen14. Moreover, the misclassification of this serovar due to the technically-demanding serotyping protocols and the evolution of multiple monophasic genotypes, make tackling the phylogenetic differentiation of MVSTm from serovar Typhimurium more challenging15,16. On the other hand, large-scale genomic approaches based on core genome multi-locus sequence typing (cgMLST) and single nucleotide polymorphisms (SNPs) phylogenies have shown an invaluable potential for WGS- subtyping of genetically closely related strain. Additionally, the constant growing of public database containing Salmonella genomes (currently including >100,000) represent an unforeseeable opportunity to analyse population structure or epidemiological transmission chain within single populations17,18. Recent studies have shown that combining core genome analysis with accessory genes pool analysis, such as pan-genome wide association studies (pan-GWAS), has improved understanding on evolutionary and phylogeographic patterns of several food-borne bacterial pathogens19,20,21,22.
Geographical structure of bacterial population is well documented for several pathogens such as Mycobacterium tuberculosis23 and Helicobacter pylori24. However, for food-borne pathogens25,26,27,28 the local phylogeographical signals quickly deteriorate by the rapid movement of lineages across the globe due to international trade of food and animals, and human travel25,29. Nevertheless, the adaptation of certain lineages to specific hosts or food production systems, which are more relevant on a local geographical scale, may result in the expansion of successful epidemic clones harbouring unique gene clusters. These clusters constitute specific biomarkers that may be used to improve source attribution in strains circulating in different countries. Therefore, in this study the phylogeographical structure of a set of 148 geographically and temporally related MVSTm isolates collected in Italy between 2012 and 2014 from human and swine was investigated in an extended context of selected publicly available S. Typhimurium/MVSTm strains from several countries.
Combining phylogenetic analysis and genome-wide association study (GWAS) we found strong evidence of the phylogeographical structure of the MVSTm isolates, identifying a specific SopEϕ-like phage as biomarker for a clone that has recently spread widely in Italy.
Results
Quality of de novo assembly
All the draft genome sequences from 148 Italian (herein STY) MVSTm isolates originating from human and swine passed the QA/QC measures as defined in INNUca pipeline (https://github.com/theInnuendoProject/INNUca). In silico MLST classified 142 (96%) samples as Sequence Type (ST)-34, 3 (2%) as ST-19, 1 (0.7%) as ST-11 and 1 (0.7%) as ST-1995. For one isolate MLST ST was not found.
Population structure Salmonella genomes
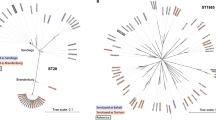
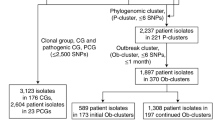
Population structure analysis has been performed based on a core genome gene-by-gene approach using the chewBBACA30 suite (https://github.com/B-UMMI/chewBBACA), to untangle the geospatial evolution of the 148 Italian STY MVSTm isolates in the context of a representative set (4,312) of publicly available Salmonella enterica genomes including the most common serovars. A total of 3,255 out of the 8,558 loci in the wgMLST schema have been detected in >99% of the samples and used for the cgMLST study. No concordance has been found between serotyping and any goeBURST clusters based on the 3,255 loci cgMLST schema (Adjusted Wallace Coefficient (AWC) <0.6). However, considering serovar 1,4,[5],12:i:- as Typhimurium, concordance between cgMLST clustering and serotyping have been found at ~30% (965) of allele differences (bidirectional AWC >0.97), including 35 goeBURST groups. At 965 cut-off, 141 out of 148 STY MVSTm isolates belong to a single group along with 2,595 genomes including the majority of S. Typhimurium and other MVSTm publicly available strains. Besides, a large part of the STY MVSTm isolates (136 out of 148; ~92%) cluster in a single goeBURST group at ~2.5% (75) of allele differences (referred as “goeBURST75”) along with a mixed population of 241 S. Typhimurium and 912 MVSTm publicly available genomes (Supplementary Table S1). The MVSTm strains were isolated between 2001 and 2017 from human (52%), swine (14%) and other sources (22%) (124 environment, 50 avian and 24 cattle). For 111 isolates no source of isolation was available. Almost the 94% of these strains were collected in Western Europe (433) and North America (427) while the remaining isolates were from Northern Europe (35), Southern Europe (10), Asia (4) and Eastern Europe (1). Publicly available MVSTm belong to ST-34 (870), ST-19 (25) ST-2379 (12), ST-2956 (1), ST-3168 (1) and ST-3224 (1). Most S. Typhimurium genomes (227) were classified as ST-34, of which more than half (115) were human isolates mainly from North America and Western Europe. For goeBURST75, the Minimum Spanning Tree (MST) based on a new cgMLST schema, including a total of 3,591 loci, has been calculated using Phyloviz 2.031 and the country of origin of the strains have been visualized on the tree (Supplementary Fig. S1). Figure S1 showed a partial enrichment of geographical linked strains in certain parts of the MST (e.g. Italian cluster, yellow circle).
Pangenome analysis
Pangenome analysis has been performed using Roary32 on a total of 1,326 genomes comprising all the 1,289 genomes belonging to goeBURST75 (including the 136 STY MVSTm isolates) and an outgroup composed by a set of 38 S. Typhimurium/MVSTm strains, including 27 Italian MVSTm, 19 of which isolated during a large outbreak in Southern Italy33. The pangenome consists of a matrix of 13,135 group of orthologues of which 9,085 are accessory (present in <99% of genomes): 8,588 present in less than 15% of strains, 333 present in more than 15% but less than 95% of strains, and 164 genes present in more than 95% of strains but less than 99%. A distance tree was inferred based on binary data of presence/absence of accessory gene using IQtree34 (Supplementary Fig. S2). Two major clusters have been identified: (a) including 1,233 strains mainly of ST-34, and (b) containing 98 strains mainly of ST-19.
Within the large ST-34 cluster, S. Typhimurium/MVSTm strains are aggregated in clades irrespectively of the year as well as of the source of isolation (Fig. S2). However, even if isolates originating from different countries are gathered together across big clades, several small clusters including isolates of the same geographical area could be visually identified across the tree. In particular, the clade named herein STY-clade (Fig. S2) is populated by 98 isolates of which roughly 71% (70) were Italian while the remainder were from Western Europe (24) and North America (4). Isolates from this clade were collected between 2007 and 2017 and obtained from human, swine, and cattle. These data suggest that certain lineages might contain specific accessory genetic markers as a result of geographical segregation.
In silico characterization of the isolates
A comprehensive list of the plasmids detected in the 1,326 genomes is available in Supplementary Table S2. The presence of plasmids has been detected in most of the genomes (1214; 91.5%) based on the positive match against PlasmidFinder database35. The most frequently reported plasmid was IncQ1 (959; 72.3%) followed by ColRNAI (506; 38.2%), Col156 (244; 18.4%). In total, the simultaneous presence of these three plasmids was observed in 77 ST-34 strains of which ~48% (37) clusters within the above described STY-clade. Overall, less than 6% of isolates were positive for further plasmid including several incompatibility groups Inc. Although none of the tested strains harboured the complete 94 kb virulent pSLT plasmid, remains of the plasmid have been detected exclusively in the ST-19 clade which includes 21 MVSTm isolates possessing from 4 to 32 of the pSLT coding DNA sequences (CDSs), comprising the virulent markers spvB and spvC36. All the MVSTm outbreak isolates characterized by Cito and colleagues33 and 2 out 6 STY MVSTm isolates clustered within ST-19 clade do not possess any pSLT CDSs. In contrast, the other 4 STY MVSTm within ST-19 clade isolates possess from 4 to 30 pSLT CDSs.
Almost all genomes (1,279; 96.4%) are positive for at least one antimicrobial resistance associated gene (ARG) and most of them (1,011; 76.2%) possess three or more ARGs, while a limited number (3.6%) did not have any positive match in Resfinder database37 (Supplementary Table S3). The most prevalent ARGs were related to resistance to tetracycline (89.7%), sulphonamides (76.6%), ampicillin (74%), streptomycin (75.6%). More specifically, the simultaneous presence of genes for resistance to ampicillin (blaTEM-1B), streptomycin (strA, strB, or aph(3“)-Ib, aph(6)-Id), sulphonamide (sul1 and sul2) and tetracycline (tet(A) or tet(B)) predicting the ASSuT resistotype (R-type) and characterizing the so called European clone have been found in 67% of the positive isolates which were originated from human and swine sources and collected between 2004 and 2017. Particularly, a total of 105 over 167 isolates of Italian origin (~83% of the genomes within the STY-clade) exhibit R-type ASSuT. Only 18 over 1,326 genomes were classified as R-type ASSuTCGTp due to the simultaneous presence of additional genes for resistance to gentamycin (aac(3)-IVa), trimethoprim (dfrA12), and chloramphenicol (cmlA1). These isolates, predicted to harbour the R-type typical of MVSTm described as part of the Spanish clone, were collected from human sources and interspersed among clusters including isolates from different sources and with R-type ASSuT.
Colistin resistance related genes mcr-1, mcr-3, mcr-4 or mcr-5 were revealed in only 10 of the ST-34 genomes mainly classified as MVSTm (8/10), collected in Italy, UK and Thailand from swine (5/10) and human. None of the genomes belonging to the STY-clade are positive for colistin resistance genes.
Genome-Wide Association Study identified genetic markers in Italian MVSTm
To investigate which genetic traits may be associated with specific MVSTm genotype that has shown a successful local expansion in Italy, we used Scoary38. Each gene cluster in the accessory genome was scored according to its apparent correlation to a predefined trait defined as Italian population, and Benjiamini-Hochberg (BH) P-value was calculated. Of the 9,085 accessory gene clusters, Scoary reported a total of 49 loci with a BH value under 0.05, and present in the 20% or more of Italian and the 30% or less of non-Italian isolates (Table 1). Loci clusters are located in separate fragments of the genomes, most of which exhibited homology to various genetic regions including phages, prophages and plasmid-associated genes originating from different bacterial species (Salmonella enterica, E. coli and Shigella). Thus, gene clusters have been divided in three groups: group 1 includes 7 contiguous loci belonging to a putative plasmid; group 2 includes 33 contiguous loci belonging to a large 42.9 kb prophage region; and group 3 includes the remaining 9 genes which are spread across the genome. These 49 loci are overrepresented in the STY-clade genomes (Supplementary Fig. S2). Particularly, only a single Italian isolate harbouring both group 1 and group 2 loci, and five UK isolates possessing a significant amount of group 2 loci are located outside the STY-clade. Finally, although clearly dominant in STY-clade, group 3 loci have been found quite frequently across the tree.
Characterization of the prophage region
A total of 10 prophages regions have been annotated by PHAST39 in the genome of the strain STY194, selected herein as reference genome for STY-clade. Of those 10 regions, 6 were intact, 3 were incomplete and one questionable. Among the intact regions, the one from nucleotide position 1,153,230 to 1,196,161 (42.9 Kb in total) includes 62 loci of which 33 have previously been classified by Scoary38 as strongly associated with Italian MVSTm (group 2, see above). The phage has been integrated downstream the tRNA-thrW gene homolog of S. Typhimurium LT2 (Fig. 1). A similar phage has been detected also in the UK strain SAL_JA6411AA (Fig. 1), belonging to the STY-clade but missing the group 2 loci. The phage in SAL_JA6411AA shows identical 3′-end but a divergent 5′-end sequence compared with STY194. The divergent part includes the 33 Italian associated loci of group 2 in STY194, substituted in SAL_JA6411AA by 22 different genes. In STY194, the 5′-half of the phage comprises genes involved in transcription and regulation, integration-recombination and cell division, prophage repression, cellular lysis and serum resistance. The conserved 3′-end of the pro-phage, which accounts for the 45.2% of the entire prophage genome, shows high similarity to the Shigella flexneri prophage SfII, and mainly encodes proteins involved in capsid formation and DNA packaging (head, tail, and terminase). Most of the 24 loci of this region were found in more than 75% of all genomes. The final portion of this phage genetic region shows homology to Salmonella phage SP_004 tail fiber assembly protein followed by sopE, a G-nucleotide exchange factor protein from SopEϕ. Genes encoding for these two proteins were shared by roughly the 35% of analysed genomes and sopE has been found to be negatively associated with North American origin (Fisher’s exact test; P < 0.0001).
MVSTm prophage region alignment. Alignment of 42.9Kb prophage region of STY194 strain (in the middle) including 62 loci of which 33 (yellow) are that classified by Scoary38 as strongly associated with Italian MVSTm (group 2 in the text). At the top, S. Typhimurium LT2 showing the insertion of the prophage between the tRNA-thrW locus downstream of proA and the transposase STM0325. At the bottom, similar prophage region of UK strain SAL_JA6411AA with divergent loci coloured in grey. The full match of shared loci (orange and magenta arrows) is showed in blue.
Phylogenomic reconstruction of the MVSTm strains
To better understand the genetic relationship of isolates characterized by different accessory genes profiles but gathered into a single goeBURST group showing up 2.5% of alleles differences, we performed a single nucleotide polymorphisms (SNPs) based phylogeny. All genome assemblies included in goeBURST75 (1,289) were mapped against the STY-clade reference genome using Snippy. Pairwise SNPs differences ranged between 0 to 1,793 with a median of 334 and a median percentage of bases aligned to the reference of the 98,43%. The maximum likelihood phylogeny and population structure were inferred based on 11,278 core SNPs. Two populations were identified corresponding to two major clades in the ML tree (Supplementary Fig. S3). Clade I is characterized by long branches and includes 25 MVSTm and one S. Typhimurium isolates of ST-19 mainly isolated in North American (~65%). More than half of the genomes exclusively included in clade I harboured from 10 to 32 pSLT-related genes. On the contrary, clade II is characterized by very short branches and includes 1,267 MVSTm (1,025) and S. Typhimurium (242) isolates mainly belonging to ST-34 (97,9%) and ASSuT genotype (66,3%), and collected in Europe (60%) and North America (38%) (Fig. 2). At 0.013 (nucleotide substitutions per site) distance from the root, clade II was divided in 61 subclades (containing at least two genomes) and 56 singletons. The subclades 10, 41 and 61, composed by 91, 195 and 474 genomes, respectively, account for ~60% of the genomes in clade II and are significantly associated to the origin of the isolates. Subclade 10 (which contains the reference genome) is significantly associated with Italian origin (Fisher’s exact test; P < 0.0001) and includes ~50% of the Italian STY-isolates available in the dataset and 5 additional Italian MVSTm isolates that were collected prior to 2012. In subclade 10, pairwise distance was from 0 to a max of 368 (Supplementary Fig. S4). In comparison to the accessory genome clustering, subclade 10 contains 85/91 isolates belonging to STY-clade and all but 5 possess the sopE-containing phage. The distribution of the loci overrepresented in the Italian isolates on the core SNP tree, as shown in Fig. 2, indicate a clear association with subclade 10, particularly for the group 1 and 2 loci. Subclade 41 is significantly associated with Western Europe origin (UK and Ireland; Fisher’s exact test; P < 0.0001) while subclade 61 is significantly associated with North American origin (Fisher’s exact test; P < 0.0001), comprising 88% of the North American isolates included in the study.
Core SNPs maximum likelihood tree. The maximum likelihood tree was inferred based on 11,278 core SNPs detected on 1,289 isolates. Figure 2 shows the tree pruned on clade II with coloured branches for subclade 10 (yellow), 41 (blue) and 61 (magenta). The internal circles indicate the originating geographical area of each isolates indicated by colours as in the legend. Externally, clusters of genes (detailed in Table 1) statistically associated with Italian strains divided by colours in plasmid-related contiguous loci (green); prophage related contiguous loci (yellow); and associated loci spread across the genome (light blue). The black hits are indicating sopE gene presence.
Bio-markers distribution
We investigated the distribution of SopEϕ containing the genes overrepresented in Italian MVSTm isolates in an extended Salmonella enterica dataset consisting of 8,787 genomes including additional 4,215 publicly available S. Typhimurium and its monophasic variant (Supplementary Table S4). The new dataset contains a total of 5,660 S. Typhimurium, 1,084 MVSTm, 1,518 Enteritidis and 525 others Salmonella serovars. All the CDSs of SopEϕ from STY194 were screened against this dataset. The presence/absence of each locus according to origin of the strain (Italian vs non-Italian) is summarized in Fig. 3, which shows a clear overrepresentation of the 5′-half of the SopEϕ in samples of Italian origin. We further divided the samples in four categories, according to percentage of positive matches of SopEϕ loci (Table 2), considering ≥90% of positive matches as positive for SopEϕ (group A). Roughly 42% (66) of the Italian MVSTm isolates over the whole dataset were included in group A along with 11 isolates from the Western European area (UK, Ireland, Luxembourg) and 2 from North America (US). Strains in group A, 74 of which are members of the Italian associated subclade 10 of goeBURST75 cluster described above, were isolated from human and swine and were collected over 10 years (2007–2017). Approximately 5% of the samples in the dataset were positive for at least 50% of the SopEϕ loci (group B), 95% of which (337) were either serovars S. Typhimurium (25%) or MVSTm (71%) and the remains belong to serovar Derby. The majority (285) of S. Typhimurium/MVSTm genomes included in group B has been collected from human samples in European countries, particularly UK (187), while a limited number (35) was from American isolates collected from different sources. Roughly 90% of the genomes belonging to the UK associated subclade 41 of goeBURST75 cluster were classified as group B. The remaining 8,356 genomes show only limited positive matches (<50%) for SopEϕ loci. Around 40% of Italian MVSTm isolates were included in group C (≥30% of loci) along with other 4,759 S. Typhimurium/MVSTm genomes collected over a century (i.e. collection timeframe ranges between 1917 and 2017) across five continents. In addition, 95% of the genomes belonging to the North America associated subclade 61 of goeBURST75 cluster were classified in group C. The remaining 1,503 S. Typhimurium/MVSTm genomes, including 4 Italian MVSTm isolates clustered in subclade 10, did show less than 30% or no positive matches (group D).
SopEϕ loci distribution between Italian and non-Italian isolates. The percentage of genomes with positive match for each locus included in sopE-containing prophage is reported in the chart according to Italian (blue) and non-Italian (orange) origin. Gene details for each locus is indicated on the left as reported by PHAST annotation.
These data suggest that this SopEϕ is rather bound up to subclade 10 of goeBURST75 cluster. We further verified that 529 out 4,217 S. Typhimurium/MVSTm genomes from the novel dataset belong to goeBURST75, including genomes classified as group A (3) and B (22). However, only genomes from group A, collected in Italy from 2007 to 2009, clustered in subclade 10 (Supplementary Fig. S5), supporting the hypothesis that this feature is characteristic of this subclade.
Discussion
The importance of S. 1,4,[5],12:i:- (MVSTm) arose when it climbed up the charts of Salmonella serovar responsible of food-borne outbreaks worldwide. In particular, in Europe the joint report on zoonosis monitoring by EFSA and ECDC described S. 1,4,[5],12:i:- as the third serovar among Salmonella already in 20136. Swine appears to be the main reservoir of this peculiar S. enterica serovar. However, the monomorphic nature of MVSTm has been an obstacle for identifying the relative importance of other animal species as sources of human infections40.
Several studies have been conducted in recent years to elucidate the phylogenetic relationship, transmission dynamics as well as the virulence and resistance key determinants of epidemics MVSTm11,12,33,41,42,43,44,45,46. Genomic analyses have suggested the emerging of multiple independent clones in the United States and Europe4. Specifically, three distinct epidemics have driven the microevolution of MVSTm across the globe resulting in the expansion of different clones which tend to be dominant in specific geographical locations41. Recent data on Salmonella serovar Cerro suggested that several genomic markers associated to geographically segregated phylogroups may contribute to the ability of Salmonella to rapidly diverge and adapt to a specific niche47. Therefore, geographical segregation can play an important role in the microevolution of emerging clones, leaving enough genetic signal in the population which can contribute to improve source attribution of clinical MVSTm infections. In the present study, we tested this hypothesis by focusing on identifying biogeographical markers in MVSTm genomes of a geographical and temporal related set of isolates obtained in Italy between 2012 and 2014. To mine phylogenetic related isolates from different part of the world, we applied a naïve approach by comparing the 148 Italian isolates with a large set (>4,000) of representative publicly available genomes of several S. enterica serovars using the gene-by-gene methodology. Thus, we identified ~1,300 S. Typhimurium and MVSTm from a broad geographical area collected in almost 20 years from several sources showing up to 2.5% allele diversity with most of the Italian isolates. By integrating phylodynamics with genome-wide association analysis, we have shown that within this peculiar population of very similar S. Typhimurium/MVSTm isolates, the expansion of genotypes in a specific geographical region is facilitated by the acquisition of unique accessory genetic markers. Phylogenetic reconstruction revealed that isolates from the same geographical origin form several highly supported monophyletic groups, providing discrete evidence of the phylogeographical structure of this population. Isolates from human and from swine related sources clustered in these groups indicating that humans are exposed to the same genotypes circulating among pigs. The presence of most of the Italian isolates collected over 7 years of sampling within a single monophyletic clade characterized by specific repertoire of plasmid- and phage-related loci supports the hypothesis that this genotype endured a substantial genetic differentiation. Among prophage elements, we found sopE gene, a virulence factor recently described in European strains41, enriched in genomes from subclades 10 and subclades 41, significantly associated with Italian and UK origin, respectively. This is consistent with findings by Petrovska and colleagues suggesting an increase of sopE gene frequency since 2007 in monophasic epidemic isolates from UK and Italy41; increase confirmed by the overrepresentation of sopE gene in the European MVSTm collected after 2010 (350/727; ~48%) analysed in this study. Although our data show a negative association of this gene with isolates from North America, a recent study by Elnekave and colleagues42 reported the presence of sopE gene in US isolates from swine samples collected during 2014–2016. The fact that MVSTm isolates harbouring sopE gene have been collected in Europe for several years before may raise the question of whether sopE positive isolates in US are most likely originating from European strains circulating in swine production chain. Further studies are needed to elucidate with higher resolution the genetic relationship of sopE positive isolates on a more representative set of US and European swine-related MVSTm strains.
Noteworthy, whereas prophage virulence gene sopE was mostly located in strains from Italian and Western Europe phylogroups in ST-34 clade II, spvC and spvB and other virulence markers of pSLT plasmid with reported contribution to pathogenicity of S. Typhimurium36,48 were located in strains from clade I belonging to ST-19. The presence of these virulence genes exclusively in ST-19 strains provided evidence that these strains most likely originated from S. Typhimurium ancestors distinct from that of the European clone. This is consistent with results of García studies13,49,50 where the presence of spvC gene and virulence-plasmids genes is reported only in MVSTm ST-19 strains. Although spvC positive ST-19 MVSTm strains have been described as similar to the hepta-resistant Spanish clone, antimicrobial resistance (AR) genes were found in only 4 out of 15 of the virulence-harbouring isolates of our dataset. In the current study, we showed that the most common AR profile for the majority of Salmonella Typhimurium/MVSTm strains isolated from humans, animals, environment and animal foodstuffs included in ST-34 clade II predict ASSuT genotype. The wide diffusion of multiple genotypes of R-type ASSuT MVSTm in European countries as well as in North America and Asia constitute a growing risk that can be associated to increased hospitalization, development of a bloodstream infection, or treatment inefficacy in patients51. However, the local expansion of specific clones can also result in the loss of AR genes, as we observed in the UK-associated subclone 41. The loss of ASSuT or ACGSSuTTp genotypes emerged towards the terminal branches of the subclade 41 tree, populated exclusively by isolates harbouring a single tetracycline resistance gene. Since within this subclade time of isolation of mono-resistant genotypes is subsequent to that of multi-resistant genotypes, we presume that the dynamic genome plasticity of S. Typhimurium/MVSTm serovars may lead to the formation and successful expansion of clones suffering the loss of particular adaptive traits.
Petrovska and colleagues41 have investigated the microevolution of MVSTm clones responsible for recent UK epidemic ways (from 2005 to 2012). The authors discovered that monophasic epidemic clones circulating in UK and Italy are characterized by the acquisition of multiple novel genes, including a sopE-containing prophage mTmV, and formed a single clade with remarkable genetic variation from North American and Spanish epidemics clones. They identified three distinct subclades one of which (i.e. subclade C) being preferentially associated with Italian livestock production. In our study, four of the Italian isolates collected up to 2010 included in the study of Petrovska and colleagues41 belong to the Italian associated subclade 10 as shown in Fig. 2, together with ~57% of the Italian STY isolates analysed in this study and collected from 2012 to 2014. Particularly, the subclade 10 represents a significant expansion of the sopE positive monophyletic group within subclade C as described by Petrovska and colleagues41. Similarly, the UK associated subclade 41, as shown in Fig. 2 herein, represents the recent expansion (>90% of the samples within subclade 41 were collected in UK or Ireland after 2014) of sopE positive monophyletic group within subclade A described by Petrovska and colleagues41. The ongoing clonal expansion of these sopE positive MVSTm subpopulations shows that the acquisition of this gene has conferred a clear competitive advantage in the ongoing European MVSTm epidemics. As previously suggested41, the acquisition of sopE has happened in multiple independent events. This theory is confirmed by the gene contents of the sopE-containing prophage. Indeed, in the UK associated subclade 41, sopE is located at the 3′-end of a prophage mTmV as previous described41, while in the Italian subclade 10 the prophage containing sopE shared only half of the mTmV genes. This novel prophage, mTmV2, contains the majority of the loci overrepresented in Italian isolates and is characteristic of subclade 10 as confirmed by investigating the prophage presence on a novel dataset of thousands of S. Typhimurium and its monophasic variant genomes. Results from this analysis underlined that no strains outside the goeBURST75 population contains mTmV2 prophage and that the addition of new genomes doesn’t change the topology of the tree, which shows mTmV2 positive genomes, classified as group A, clustering within subclade 10.
The gain and loss of mobile genetic elements may “unlock the secrets” for the optimization of infection-control strategies and effective containment of emergent pathogens, as was already discussed in a recent study on the transmission dynamics of Enterococcus faecium52. Therefore, we can conclude that investigating on the presence of particular genetic elements, such as mTmV2 prophage, can contribute in enhancing the ability in tracking the dissemination of specific clones of MVSTm in ongoing epidemics.
Methods
Bacterial strains, genome sequencing and assembling
A total of 148 Salmonella enterica serovar Typhimurium variant 4,[5],12:i:- (MVSTm) have been collected from different Italian regions between 2012 and 2014 during a surveillance study. For the aim of this study, this dataset has been named STY. Pig faecal samples (11), pork carcass isolates (23) and pork meat at retail isolates (27) were obtained from the Italian National Reference Laboratory for Salmonella (NRL Salmonella, Istituto Zooprofilattico Sperimentale delle Venezie, Legnaro, Italy), while isolates from humans with gastroenteritis (82) were obtained from the National Institute of Health (Instituto Superiore della Sanitá, Roma). Genomic DNA of the 148 STY isolates was extracted and purified using the HWD DNA minikit (QIAGEN) according to the manufacturer’s instruction. Index-tagged paired-end Illumina sequencing libraries were prepared using NexteraXT library preparation kit and whole genome sequencing was performed on Illumina MiSeq platform generating tagged 250 bp paired-end reads. The paired-end raw reads were assembled using the INNUca pipeline53 (https://github.com/theInnuendoProject/INNUca), which consists of several modules and QA/QC steps. In brief, INNUca starts by calculating if the sample raw data fulfil the expected coverage (min 15x). After subjecting reads to quality analysis using FastQC, and cleaning with Trimmomatic54, INNUca proceeds to de novo draft genome assembly with SPAdes55 v3.11 checking assembly depth of coverage (min 30x). Finally, Pilon56 improves the draft genome by correcting bases, fixing misassembles and filling gaps; prior species confirmation and seven genes MLST Sequence Type (ST) is assigned with mlst software (https://github.com/tseemann/mlst).
Reference genomic collection for investigating Salmonella population structure
To assess the population structure of monophasic Salmonella Typhimurium isolates in comparison to a Salmonella enterica reference collection, 4,312 publicly available draft or complete genome assemblies along with available metadata have been downloaded from public repositories (i.e. EnteroBase - https://enterobase.warwick.ac.uk/, National Center for Biotechnology Information NCBI - https://www.ncbi.nlm.nih.gov/ and The European Bioinformatics Institute EMBL-EBI - https://www.ebi.ac.uk/; accessed April 2017). The reference collection includes 1,465 Salmonella enterica ser. Enteritidis, 1,425 ser. Typhimurium, 985 MVSTm, and 437 of other frequently isolated serovars in Europe according to EFSA and ECDC joint summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks (EFSA/ECDC, 2016). All available assemblies for MVSTm have been chosen. For each of the other serovars, genomes have been selected to maintain the same proportions of genetic diversity representatives of all the diversity revealed by rMLST18 as existing in EnteroBase at the date of collection (April 2017) and to design a suitable and accurate cgMLST schema which was also able to validate if the genomes serotypes were correctly annotated in the available metadata.
Core genome MLST (cgMLST) allele calling and cluster analysis
Population structure analysis of Salmonella genomes have been performed using cgMLST methodology57. Schema curation, validation and allele calling have been carried out using the chewBBACA58 suite as described by Rossi and colleagues59 (https://github.com/theInnendoProject/chewBBACA_schemas). Briefly, the wgMLST schema V2 from EnteroBase, including 21,064 loci, have been downloaded and curated using chewBBACA suite resulting in a total of 8,558 loci. The core genome MLST profile, defined as the loci presence in at least the 99% of the samples, has been then extracted using chewBBACA ExtractCgMLST consisting of 3,255 loci. Genomes with more than 2% of missing loci have been excluded.
The global optimal eBUSRT algorithm (goeBURST)60 implemented in Phyloviz31 2.0 has been used to identify cluster membership of cgMLST profiles of Salmonella strains at different thresholds of allelic differences. Neighbourhood Adjusted Wallace Coefficient (nAWC)61 was calculated to assess cluster consolidation dynamics. nAWC examines the congruence of partitions between adjacent similarity thresholds used for cluster definition (https://github.com/theInnuendoProject/nAWC). nAWC identifies areas in in which distance thresholds produce robust clusters reflecting basic units in Salmonella overall population structure61. In addition, we also evaluate concordance between partitions obtained at different goeBURST cut-off and serotyping with Adjusted Wallace Coefficient62 (AWC).
Genome annotation, pangenome analyses and Genome-Wide Association Study (GWAS)
Pangenome analysis was performed on all genomes belonging to a selected goeBURST group, henceforth referred to as goeBURST75, previously defined for showing up to 2.5% of allele differences and for including 136 newly sequenced STY MVSTm isolates. A set of outliers composed of 38 S. Typhimurium/MVSTm genomes, including additional 27 Italian MVSTm outbreak-related strains, was also included in the analysis in order to gain further genome diversity and comparability. Salmonella genomes were annotated with Prokka63 (https://github.com/tseemann/prokka) and the produced GFF3 files were used to generate the pan-genome matrix with Roary32 (https://github.com/sanger-pathogens/Roary) using default parameters. The neighbourhood joining tree based on the binary matrix of presence and absence of accessory genes calculated by Roary32 was visualized on iTOL64 v3 (https://itol.embl.de) along with relevant metadata. A GWAS was performed based on Roary32 results using Scoary38 (https://github.com/AdmiralenOla/Scoary) v1.6.16 with default parameters. Patterns of genes were reported as significantly associated to geographical origin (e.g. Italy) if they attained a Benjiamini-Hochberg-corrected P-value less than 0.05 and were present in at least the 20% of the selected isolates (e.g. Italian) and absent in at least the 70% of the rest of the dataset (e.g. non-Italian isolates). The synteny of the associated loci was visually assessed using Artemis65 annotation tool on a selection of STY isolates and further manually annotated. If the associated genes were annotated as hypothetical protein the gene was manually curated by searching homologs sequencing in non-redundant (nr) NCBI protein sequences collection using blast+66 v2.7.1 (https://blast.ncbi.nlm.nih.gov/). We investigated presence and distribution of the geographic associated loci on an extended Salmonella enterica dataset inclusive of 8,787 genomes (Supplementary Table S4). This dataset included all genomes of the reference collection along with a novel curated set of 4,217 publicly available S. Typhimurium genomes: all public genomes not already included in the reference collection, deposed in SRA or ENA as S. Typhimurium or its monophasic variant for which country of isolation was available and of which assembly met minimum quality criteria (i.e. genome assembly length ranging 4.5–5.5 Mb). Presence/absence of the associated loci were performed using chewBBACA workflow58. Briefly, a schema composed by the associated loci were constructed and used for performing allele calling on all 8,787 genomes. The locus was marked as present if an allele was called or if it was annotated as “non-informative paralogous hit (NIPH/NIPHEM)” or if was detected on the tip of the query genome contigs58 (for details https://github.com/B-UMMI/chewBBACA/wiki/2.-Allele-Calling).
In silico typing
Antibiotic resistance and plasmids prediction was performed with ABRicate pipeline (https://github.com/tseemann/abricate) using ResFinder37 and PladmidFinder35 as reference database. The typical AR profile of S. 1,4,[5],12:i:- “European clone” was defined as the simultaneous presence of blaTEM-1, strA (and its synonymous aph(3“)-Ib), strB (and its synonymous aph(6)-Id), sul1, sul2 and tet(B) genes (R-type ASSuT)12. In addition, when cmlA1, aac(3)-IV and dfrA12 genes were detected in that isolates harbouring ASSuT related genes they were predicted as resistance type ASSuTCGTp, a specific pattern associated to S. 4,[5],12:i:- from the “Spanish clone”13.
The presence of pSLT-genes encoding virulence factor in a 94-kb plasmid (AE006471) from S. Typhimurium LT2 was investigated using blastn implemented in blast+66 v2.7.1. The PHAge Search Tool (PHAST)39 was used to identify the positions of putative phage elements. For PHAST analysis, genomes have been annotated using RAST annotation server67. Hence, the annotated GBK file was uploaded to the public PHAST web server (http://phast.wishartlab.com/)39.
Single-nucleotide polymorphism (SNP) analysis
To establish the phylogenetic relationship between closely related strains based on the goeBURST clustering, SNP analysis have been performed with Snippy v3.2 pipeline, using the assembled genomes as input files (https://github.com/tseemann/snippy). As reference, the best assembled draft genome (based on N-50 values and coverage) harbouring the largest set of geographical associated genes has been selected within the goeBURST75 cluster. A core alignment of all the conserved nucleotide variant sites present in all genomes was used to build a maximum-likelihood tree using IQ-tree34 with a gamma correction for site rate variation using 1,000 bootstrap replicates to support the nodes. hierBAPS68 was used for clustering the samples based on the core SNPs alignment up to second level.
Data Availability
Genome assemblies are accessible at the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the project accession number: PRJEB23875.
References
Switt, A. I. M., Soyer, Y., Warnick, L. D. & Wiedmann, M. Emergence, Distribution, and Molecular and Phenotypic Characteristics of Salmonella enterica Serotype 4,5,12:i:–. Foodborne Pathog. Dis. 6, 407–415 (2009).
Hopkins, K. L. et al. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 15, 19580 (2010).
Echeita, M. A., Aladueña, A., Cruchaga, S. & Usera, M. A. Emergence and Spread of an Atypical Salmonella enterica subsp. enterica Serotype 4,5,12:i:− Strain in Spain. J. Clin. Microbiol. 37, 3425–3425 (1999).
Soyer, Y. et al. Salmonella enterica Serotype 4,5,12:i:−, an Emerging Salmonella Serotype That Represents Multiple Distinct Clones. J. Clin. Microbiol. 47, 3546–3556 (2009).
Zamperini, K. et al. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:- from poultry is a variant Typhimurium serovar. Avian Dis. 51, 958–964 (2007).
European Food Safety Authority & European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 14, n/a–n/a (2016).
Mossong, J. et al. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:i:- in Luxembourg, 2006. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 12, E11–12 (2007).
Dionisi, A. M. et al. Molecular Characterization of Multidrug-Resistant Strains of Salmonella enterica Serotype Typhimurium and Monophasic Variant (S. 4,[5],12:i:–) Isolated from Human Infections in Italy. Foodborne Pathog. Dis. 6, 711–717 (2009).
Hauser, E. et al. Pork Contaminated with Salmonella enterica Serovar 4,[5],12:i:−, an Emerging Health Risk for Humans. Appl. Environ. Microbiol. 76, 4601–4610 (2010).
Echeita, M. A., Herrera, S. & Usera, M. A. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39, 2981–2983 (2001).
Ido, N. et al. Characteristics of Salmonella enterica Serovar 4,[5],12:i:- as a Monophasic Variant of Serovar Typhimurium. PLoS ONE 9 (2014).
García, P. et al. Horizontal Acquisition of a Multidrug-Resistance Module (R-type ASSuT) Is Responsible for the Monophasic Phenotype in a Widespread Clone of Salmonella Serovar 4,[5],12:i:-. Front. Microbiol. 7 (2016).
García, P., Malorny, B., Hauser, E., Mendoza, M. C. & Rodicio, M. R. Genetic Types, Gene Repertoire, and Evolution of Isolates of the Salmonella enterica Serovar 4,5,12:i:− Spanish Clone Assigned to Different Phage Types. J. Clin. Microbiol. 51, 973–978 (2013).
Achtman, M. Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Philos. Trans. R. Soc. B Biol. Sci. 367, 860 (2012).
Hopkins, K. L., de Pinna, E. & Wain, J. Prevalence of Salmonella enterica serovar 4,[5],12:i:- in England and Wales, 2010. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 17 (2012).
Wain, J. & O’Grady, J. Genomic Diversity in Salmonella enterica. In Applied Genomics of Foodborne Pathogens 91–107, https://doi.org/10.1007/978-3-319-43751-4_6 (Springer, Cham, 2017).
Taboada, E. N., Graham, M. R., Carriço, J. A. & Van Domselaar, G. Food Safety in the Age of Next Generation Sequencing, Bioinformatics, and Open Data Access. Front. Microbiol. 8 (2017).
Alikhan, N.-F., Zhou, Z., Sergeant, M. J. & Achtman, M. A genomic overview of the population structure of Salmonella. PLOS Genet. 14, e1007261 (2018).
Holt, K. E. et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 112, E3574–E3581 (2015).
Skallerup, P., Espinosa-Gongora, C., Jørgensen, C. B., Guardabassi, L. & Fredholm, M. Genome-wide association study reveals a locus for nasal carriage of Staphylococcus aureus in Danish crossbred pigs. BMC Vet. Res. 11 (2015).
Reuter, S. et al. Directional gene flow and ecological separation in Yersinia enterocolitica. Microb. Genomics 1 (2015).
Bazinet, A. L. Pan-genome and phylogeny of Bacillus cereus sensu lato. BMC Evol. Biol. 17 (2017).
Achtman, M. Evolution, Population Structure, and Phylogeography of Genetically Monomorphic Bacterial Pathogens. Annu. Rev. Microbiol. 62, 53–70 (2008).
Moodley, Y. & Linz, B. Helicobacter pylori Sequences Reflect Past Human Migrations. Genome Dyn. 6, 62–74 (2009).
Llarena, A. K. et al. Monomorphic genotypes within a generalist lineage of Campylobacter jejuni show signs of global dispersion. Microb. Genomics 2 (2016).
The, H. C., Thanh, D. P., Holt, K. E., Thomson, N. R. & Baker, S. The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat. Rev. Microbiol. 14, (235 (2016).
Strachan, N. J. C. et al. Whole Genome Sequencing demonstrates that Geographic Variation of Escherichia coli O157 Genotypes Dominates Host Association. Sci. Rep. 5 (2015).
Wong, V. K. et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat. Genet. 47, 632–639 (2015).
Pascoe, B. et al. Local genes for local bacteria: Evidence of allopatry in the genomes of transatlantic Campylobacter populations. Mol. Ecol. 26, 4497–4508 (2017).
Silva, M. et al. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. bioRxiv 173146, https://doi.org/10.1101/173146 (2017).
Nascimento, M. et al. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinforma. Oxf. Engl. 33, 128–129 (2017).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Cito, F. et al. Outbreak of unusual Salmonella enterica serovar Typhimurium monophasic variant 1,4 [5],12:i:-, Italy, June 2013 to September 2014. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 21 (2016).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Carattoli, A. et al. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Gulig, P. A. et al. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol. 7, 825–830 (1993).
Kleinheinz, K. A., Joensen, K. G. & Larsen, M. V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4 (2014).
Brynildsrud, O., Bohlin, J., Scheffer, L. & Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 17, 238 (2016).
Zhou, Y., Liang, Y., Lynch, K. H., Dennis, J. J. & Wishart, D. S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 39, W347–W352 (2011).
Barco, L. et al. Ascertaining the relationship between Salmonella Typhimurium and Salmonella 4,[5],12:i:- by MLVA and inferring the sources of human salmonellosis due to the two serovars in Italy. Front. Microbiol. 6 (2015).
Petrovska, L. et al. Microevolution during the emergence of a monophasic Salmonella Typhimurium epidemic in the United Kingdom. Emerg. Infect. Dis. 22 (2016).
Elnekave, E. et al. Salmonella enterica serotype 4,[5],12:i:- in swine in the United States Midwest: an emerging multidrug resistant clone. Clin. Infect. Dis., https://doi.org/10.1093/cid/cix909 (2017).
Yang, X. et al. Prevalence and Characterization of Monophasic Salmonella Serovar 1,4,[5],12:i:- of Food Origin in China. PLOS ONE 10, e0137967 (2015).
Andrés-Barranco, S., Vico, J. P., Marín, C. M., Herrera-León, S. & Mainar-Jaime, R. C. Characterization of Salmonella enterica Serovar Typhimurium Isolates from Pigs and Pig Environment-Related Sources and Evidence of New Circulating Monophasic Strains in Spain. J. Food Prot. 79, 407–412 (2016).
Seixas, R. et al. Phenotypic and Molecular Characterization of Salmonella 1,4,[5],12:i:- R-Type ASSuT Isolates from Humans, Animals, and Environment in Portugal, 2006–2011. Foodborne Pathog. Dis. 13, 633–641 (2016).
Gymoese, P. et al. Investigation of Outbreaks of Salmonella enterica Serovar Typhimurium and Its Monophasic Variants Using Whole-Genome Sequencing, Denmark. Emerg. Infect. Dis. 23, 1631–1639 (2017).
Kovac, J. et al. Temporal Genomic Phylogeny Reconstruction Indicates a Geospatial Transmission Path of Salmonella Cerro in the United States and a Clade-Specific Loss of Hydrogen Sulfide Production. Front. Microbiol. 8 (2017).
Fàbrega, A. & Vila, J. Salmonella enterica Serovar Typhimurium Skills To Succeed in the Host: Virulence and Regulation. Clin. Microbiol. Rev. 26, 308–341 (2013).
García, P., Guerra, B., Bances, M., Mendoza, M. C. & Rodicio, M. R. IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging Salmonella enterica serotype 4,[5],12:i: −. J. Antimicrob. Chemother. 66, 543–549 (2011).
García, P. et al. Diversity of Plasmids Encoding Virulence and Resistance Functions in Salmonella enterica subsp. enterica Serovar Typhimurium Monophasic Variant 4,[5],12:i:- Strains Circulating in Europe. PLoS ONE 9 (2014).
Centres for Disease Control and Prevention. Multistate Outbreak of Multidrug-Resistant Salmonella I 4,[5],12:i:- or Salmonella Infantis Infections Linked to Pork (Final Update) Available at, https://www.cdc.gov/salmonella/pork-08-15/index.html (2015).
van Hal, S. J. et al. Evolutionary dynamics of Enterococcus faecium reveals complex genomic relationships between isolates with independent emergence of vancomycin resistance. Microb. Genomics 2 (2016).
Machado, M. et al. GitHub - B-UMMI/INNUca: INNUENDO quality control of reads, de novo assembly and contigs quality assessment, and possible contamination search. Available at: https://github.com/theInnuendoProject/INNUca.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 19, 455–477 (2012).
Walker, B. J. et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLOS ONE 9, e112963 (2014).
Maiden, M. C. J. et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736 (2013).
Silva, M. et al. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genomics (2018).
Rossi, M. et al. INNUENDO whole and core genome MLST databases and schemas for foodborne pathogens. chewBBACA_schemas: wgMLST schemas formatted for chewBBACA for food-borne pathogens. (2018).
Francisco, A. P. et al. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13, 87 (2012).
Barker, D. O. R. et al. Rapid Identification of Stable Clusters in Bacterial Populations Using the Adjusted Wallace Coefficient. bioRxiv. Available at: https://www.biorxiv.org/content/early/2018/04/16/299347.
Severiano, A., Pinto, F. R., Ramirez, M. & Carriço, J. A. Adjusted Wallace Coefficient as a Measure of Congruence between Typing Methods ∇. J. Clin. Microbiol. 49, 3997–4000 (2011).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 30, 2068–2069 (2014).
Letunic, I. & Bork, P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–245 (2016).
Carver, T. et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676 (2008).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Cheng, L., Connor, T. R., Sirén, J., Aanensen, D. M. & Corander, J. Hierarchical and Spatially Explicit Clustering of DNA Sequences with BAPS Software. Mol. Biol. Evol. 30, 1224–1228 (2013).
Acknowledgements
This study has been supported by COMPARE project (https://www.compare-europe.eu) co-funded by the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 643476 (“Collaborative management platform for detection and analyses of (re-) emerging and foodborne outbreaks in Europe”). Study has also been supported by INNUENDO project (https://www.innuendoweb.org) co-funded by the European Food Safety Authority (EFSA), grant agreement GP/EFSA/AFSCO/2015/01/CT2 (“New approaches in identifying and characterizing microbial and chemical hazards”). The conclusions, findings, and opinions expressed in this review paper reflect only the view of the authors and not the official position of the European Food Safety Authority (EFSA). The authors wish to thank CSC- Tieteen tietotekniikan keskus Oy for providing access to cloud computing resources.
Author information
Authors and Affiliations
Contributions
M.R. designed the study and draft the manuscript. F.P. performed data analyses, prepared figures and wrote the manuscript. M.S. developed the chewBBACA pipeline and collect the publicly available dataset. A.P. whole genome sequenced bacterial isolates. D.O.R.B. and E.N.T. collected the publicly available dataset and revised the manuscript. Fr.P. and G.M. collected the samples and revised the manuscript. G.M. coordinated the study. All authors have contributed to data interpretation, have critically reviewed the manuscript, and approved the final version as submitted.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Palma, F., Manfreda, G., Silva, M. et al. Genome-wide identification of geographical segregated genetic markers in Salmonella enterica serovar Typhimurium variant 4,[5],12:i:-. Sci Rep 8, 15251 (2018). https://doi.org/10.1038/s41598-018-33266-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-33266-5
Keywords
This article is cited by
-
Relationship and distribution of Salmonella enterica serovar I 4,[5],12:i:- strain sequences in the NCBI Pathogen Detection database
BMC Genomics (2022)
-
Genome-based Salmonella serotyping as the new gold standard
Scientific Reports (2020)
-
Genetic and metabolic signatures of Salmonella enterica subsp. enterica associated with animal sources at the pangenomic scale
BMC Genomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.