Abstract
Both phytohormones and non-coding microRNAs (miRNAs) play important role in root development in Arabidopsis thaliana. Mature miR166/165 s, which are derived from precursor transcripts of concerned genes, regulate developmental processes, including leaf and root patterning, by targeting Class III HOMEODOMAIN LEUCINE-ZIPPER (HD-ZIP III) transcription factors (TFs). However, their regulation through hormones remained poorly understood. Here, we show that several phytohormones dynamically regulate the spatio-temporal expression pattern of miR166/165 and target HD-ZIP IIIs in developing roots. Hormone signaling pathway mutants show differential expression pattern of miR166/165, providing further genetic evidence for multilayered regulation of these genes through phytohormones. We further show that a crosstalk of at least six different phytohormones regulate the miR166/165, their target HD-ZIP IIIs, and KANADI (KANs). Our results suggest that HD-ZIP IIIs mediated root development is modulated both transcriptionally through phytohormones and KANs, and post-transcriptionally by miR166/165 that in turn are also regulated by the phytohormonal crosstalk.
Similar content being viewed by others
Introduction
Phytohormones and several TFs play major role in root growth and development in Arabidopsis. Among these factors, several growth regulators known as phytohormones play important role during root growth and development1, 2. Phytohormones, such as auxin (Indole-3-acetic acid, IAA), gibberellic acid (GA), cytokinins (CKs), abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), brassinosteroid (BR), etc. regulate various plant developmental processes by modulating the expression of downstream genes. Auxin is known as the master regulator of plant development, as it controls almost every developmental process in plants, including root development3, 4. Apart from auxin, GA also controls root growth by regulating cell elongation and proliferation through DELLA protein degradation5. CK acts antagonistically to auxin in root development by modulating the expression of auxin efflux carriers PIN-FORMED (PINs)6. ABA negatively regulates both primary and lateral root (LR) development7. Exogenous application of JA inhibits primary root growth at all concentrations, but induces LR formation at low concentrations whereas reduces at higher concentration8. SA also regulates root development in Arabidopsis 9. Root development involves gene regulation by a complex crosstalk among different hormone signaling pathways10.
Besides role in other developmental processes, several miRNAs have also been reported to regulate root development in Arabidopsis 11. An evidence of link between phytohormones and miRNAs was identified by the study of hyponastic leaves1 (hyl1) mutant, which is defective in miRNA biogenesis and display abnormal response to different phytohormones such as ABA, auxin, and CK12.
Spatio-temporal expression pattern of genes largely contributes to their developmental role. Spatio-temporal expression or transcriptional pattern of different miRNA species of a family as well as their targets depends on the cis-regulatory elements, including hormone response elements, in their respective promoters. Therefore, to understand miRNA function it is necessary to decipher their upstream regulation. miR165 and miR166 are the two miRNAs that differ by only one nucleotide in their mature sequence and both target transcripts of same HD-ZIP III gene family members13. Arabidopsis genome encodes seven MIR166 (MIR166A–G) and two MIR165 (MIR165A-B), which finally produce same mature miR166 and miR165, respectively. miR166/165 regulates diverse developmental processes including shoot apical meristem (SAM) maintenance, leaf polarity, floral development, and root development in plants by negatively regulating HD-ZIP III gene family members - PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ARABIDOPSIS THALIANA HOMEOBOX 8 (ATHB8) and ARABIDOPSIS THALIANA HOMEOBOX 15 (ATHB15)14,15,16,17. Recently, miR166/165 has been shown to be involved in root development in Arabidopsis 18, 19. KANADI genes (KAN1 - KAN4), members of GARP transcription factors family show expression pattern complementary to HD-ZIP IIIs and also regulate HD-ZIP IIIs expression and function20. Genetic analyses revealed that HD-ZIP IIIs and KAN genes act antagonistically to each other to regulate organ patterning21, 22. Both miR166/165 and KAN genes are involved in maintaining abaxial fate of lateral organs by restricting the expression of HD-ZIP IIIs in the adaxial domain21. Although mature miR166 derived from all the seven MIR166 genes are same, each individual MIR166 gene exhibits specific spatio-temporal regulation14. Thus, the precise spatio-temporal expression of seven MIR166 genes is possibly controlled by their transcriptional regulation in specific cell-types at specific stages of development.
Spatio-temporally regulated expression dynamics of a miRNA gene family is likely to regulate the expression pattern or accumulation of their target transcripts at post-transcriptional level. We have previously shown that promoter sequences of seven MIR166 genes are significantly dissimilar to each other indicating the variations in the cis-regulatory elements in their promoters23. In the present study, we have analyzed the effects of various phytohormones on the expression pattern of miR166/165 s (precursors and mature), their target HD-ZIP IIIs and KAN genes during root development. We here show that various phytohormones influence the spatio-temporal expression pattern of all these genes, which often is due to their transcriptional regulation. We provide evidence that it is the dose of functional HD-ZIP III transcripts, regulated both at transcriptional and post-transcriptional level, which plays important role in root development. Our results uncover a complex hormonal crosstalk regulating HD-ZIP IIIs transcript level (at transcriptional/post-transcriptional level) by modulating the expression of miR166/165, KANs and hormone signaling during root development.
Results
Phytohormones dynamically regulate the expression pattern of miR166/165 in roots
Phytohormones such as auxin (IAA), GA, CK, JA, etc. regulate various aspects of plant growth and development24. Before analyzing hormonal regulation of miR166/165, we performed the growth analysis of wild type (Col-0) Arabidopsis roots in response to six phytohormones - IAA, GA, BAP, ABA, JA, and SA to test the reproducibility of phenotype in our growth conditions (Supplemental Fig. S1). Consistent with previous reports, our results confirmed that root growth and development are affected by aforesaid phytohormones.
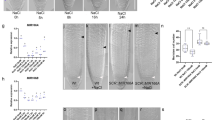
To understand whether phytohormones regulate the expression of miR166/165 and contribute to miR166/165 mediated root development, we analyzed the expression of miR166/165 upon treatment with aforesaid phytohormones at different time intervals (1, 6, 12, and 24 hrs). Treatment with 10 µM IAA significantly reduced the level of mature miR166/165 at each time point, except at 1 hr of the study, as observed in stem loop qRT-PCR analysis (Fig. 1A). Whole mount in situ hybridization experiment illustrated the reduced accumulation of miR166/165 in IAA treated roots than control roots (Fig. 1G,H). GA treatment induced the expression level of miR166/165 at all time points, showing highest upregulation up to 2.5 fold at 6 hrs of treatment (Fig. 1B). After 12 and 24 hrs of GA treatment, the expression level of miR166/165 remained higher than the untreated control but lower than that of 6 hrs of the treatment (Fig. 1B). However, the whole mount in situ localization experiment illustrated the increased accumulation of miR166/165 after the treatment with GA at 12 hrs (Supplemental Fig. S3A,C). Treatment with 6-Benzylaminopurine (BAP or CK) induced the expression level of miR166/165 at each time points, except at 1 hr, where miR166/165 level was reduced in comparison to the wild type. After 1 hr, the expression of miR166/165 was induced in BAP treatment, whereas highest increase upto 5 fold was observed at 6 hrs of treatment (Fig. 1D). The whole mount in situ localization experiment showed the increased accumulation of miR166/165 after the treatment with BAP at 12 hrs (Supplemental Fig. S3A,B). ABA treatment led to no significant change in the expression of miR166/165 after 1 hr. After 6, 12, and 24 hrs of ABA treatment, the expression level of mature miR166/165 was upregulated by 2.2, 1.5 and 37–40 folds, respectively (Fig. 1C). Whole mount in situ localization experiment also confirmed the stem-loop qRT-PCR results exhibiting increased accumulation of miR166/165 upon ABA treatment (Fig. 1G,I). JA treated roots showed increased expression of mature miR166/165 at 1, 6, 12, and 24 hrs of the treatment (Fig. 1E). Upon SA treatment, the expression of mature miR166/165 was induced at 1, 6 and 12 hrs. However, the expression of miR166/165 was downregulated after 24 hrs of the treatment (Fig. 1F). The whole mount in situ localization showed increased accumulation of miR166/165 in 7 dag roots treated with JA for 12 hrs in comparison to control (Fig. 1G,J). Localization of miR166/165 in SA treated 7 dag roots at 12 hrs showed slight difference in expression pattern to that of qRT-PCR (Supplemental Fig. S3A,D). We cannot rule out that this difference might be due to fluctuation and stability of mature miR166/165 among treatment points and/or the sensitivity of two techniques. The tissue used during qRT-PCR includes whole root (both primary root and LR) which may lead to the change in expression pattern between qRT-PCR and whole mount in situ. These results suggest that phytohormones dynamically regulate the expression of miR166/165 during stages of root development.
Phytohormones affect the expression level of mature miR166/165. The expression level of miR166/165 was quantified using stem-loop qRT-PCR in roots at 1, 6, 12, and 24 hrs after treatment with (A) 10 µM IAA, (B) 10 µM GA, (C) 10 µM ABA, (D) 10 µM BAP, (E) 20 µM JA, and (F) 100 µM SA. Whole mount in situ localization of miR166/165 was performed in 7 dag roots following 12 hrs of phytohormone treatment (G–J). (G) Control (without any treatment), (H) 10 µM IAA, (I) 10 µM ABA and with (J) 20 µM JA. In situ localization of miR166/165 in BAP, GA and SA treatment has been provided in Supplemental Fig. S3. Error bars indicate ± SE of two independent biological experiments. One-way ANOVA was performed. Statistically significant differences are indicated as, * for P < 0.05, ** for P < 0.01 and *** for P < 0.001. Scale bar indicates 50 µm.
Differential expression of MIR166 precursors contributes to the altered level of miR166/165 level in response to hormones
We observed that phytohormones affect the expression of mature miR166/165 and root development. We analyzed the effect of hormone treatment on transcriptional regulation of MIR166A–G genes at different time points (6, 12 and 24 hrs) by qRT-PCR (Fig. 2). Interestingly, exogenous application of 10 µM IAA led to the downregulation of MIR166A–G genes at all three time points (Fig. 2A). IAA treatment produced similar effect on the expression pattern of both mature miR166 and its precursors. Exogenous application of 10 µM GA induced the expression of MIR166B after 6 hrs of the treatment (Fig. 2B). We observed significant induction in the transcript levels of all MIR166 genes, except MIR166G, after 12 hrs of treatment (Fig. 2B). The expression of all MIR166 precursors except MIR166A and MIR166C was further induced by up to 1.5 folds and above at 24 hrs of GA treatment (Fig. 2B). Exogenous application of 10 µM BAP induced the expression of all MIR166 members, except MIR166D after 6 hrs of treatment. After 12 hrs of BAP treatment, MIR166A, C, E and F were upregulated whereas MIR166B, D and G were downregulated (Fig. 2C). The expression level of MIR166A–G was induced at 24 hrs of treatment (Fig. 2C). Treatment with 10 µM ABA induced the expression of all precursors by 2 to 15 folds as compared to the control, immediately after 6 hrs of the treatment (Fig. 2D). At 12 hrs of the ABA treatment, MIR166D and MIR166G showed increased expression, MIR166A expression remained unchanged and MIR166B, C, F, G expression levels were reduced in comparison to untreated control (Fig. 2D). At 24 hrs of ABA treatment, the expression of MIR166B-G decreased to almost undetectable level, except for MIR166A. The application of 20 µM JA and 100 µM SA induced the expression of MIR166A-F except MIR166G at 6 hrs of the treatment, which was detected below that of untreated control (Fig. 2E,F). The transcript level of all precursors further increased as compared to the untreated control at 12 hrs of JA and SA treatment (Fig. 2E,F). Finally, the expression of all the precursors was reduced below the untreated control at 24 hrs of JA and SA treatment (Fig. 2E,F). These differential expression patterns of seven MIR166 precursors at different time intervals of treatment indicated their temporal transcriptional regulation by developmentally important phytohormones.
Phytohormones affect the expression level of precursor genes of MIR166. The expression level of seven MIR166(A–G) genes was quantified using qRT-PCR in roots at 1, 6, 12, and 24 hrs with hormone concentrations of (A) 10 µM IAA, (B) 10 µM GA, (C) 10 µM BAP, (D) 10 µM ABA, (E) 20 µM JA and (F) 100 µM SA. The control indicates without treatment in each case. Error bars indicate ± SE of two independent biological experiments. One-way ANOVA was performed. Statistically significant differences are indicated as, * for P < 0.05,** for P < 0.01 and *** for P < 0.001.
Mutants defective in hormone signaling show altered expression of miR166/165
To confirm the transcriptional regulation of miR166/165 through phytohormones, we analyzed the expression of miR166/165 in hormone insensitive mutants. Stem-loop qRT-PCR results showed that the exogenous IAA treatment led to the downregulation of miR166/165 expression level. To further confirm our result, we checked the expression of miR166/165 in seedlings of IAA insensitive auxin resistant2-1 (axr2-1) mutant, which is defective in auxin perception and root development25. Interestingly, we observed that axr2-1 mutant root had significantly reduced level of miR166/165 expression (Fig. 3A). The reduced expression of miR166/165 in axr2-1 could be the result of increased endogenous auxin in axr2-1, as observed in case of shy2-2 26.
The expression of miR166/165 was affected in hormone insensitive mutants. The expression level of miR166/165 was quantified using stem-loop qRT-PCR in (A) axr2-1 mutant. (B) GA insensitive pentuple (gai-t6 rga-t2 rgl1-1 rgl2-1 rgl3-1) mutant. (C) aba1-1 mutant. (D) arr1-3 arr12-1 mutant. (E) coi1-16 ein2-1 pen2-4 mutant. (F) npr1-2 mutant. Error bars indicate ± SE of two independent biological experiments. One-way ANOVA was performed. Statistically significant differences are indicated as, * for P < 0.05,** for P < 0.01 and *** for P < 0.001.
Stem-loop qRT-PCR results showed that the exogenous GA treatment led to the upregulation of miR166/165 level. To further confirm our result, we checked the expression of miR166/165 in roots of GA insensitive pentuple mutant of all five DELLA proteins, gai-t6 rga-t2 rgl1-1rgl2-1rgl3-1 using stem-loop qRT-PCR analysis. When we checked the expression of miR166/165 in DELLA pentuple mutant, we found increased expression level of miR166/165, similar to the exogenous GA treatment (Fig. 3B).
The exogenous ABA application inhibited primary root growth (Supplemental Fig. S1C). We observed significant induction of miR166/165 expression by ABA treatment in comparison to wild type. To confirm genetically, we further checked the expression of miR166/165 in aba1-1 mutant, an ABA biosynthetic mutant, which is ABA deficient27. Roots of aba1-1 mutant showed reduced expression level of miR166/165 in comparison to wild type (Fig. 3C).
To confirm cytokinin mediated regulation of miR166/165, we checked the expression of miR166/165 in arr1-3 arr12-1 double mutant, which shows CK insensitivity towards root elongation, LR formation and callus induction28. Since exogenous CK inhibits primary root elongation, arr1 arr12 mutant shows longer root phenotype suggesting CK insensitivity29. In our experiment, exogenous BAP treatment induced the expression of miR166/165. In comparison to wild type, the expression of miR166/165 was reduced in arr1 arr12 mutant, which further confirmed the CK mediated regulation of miR166/165 (Fig. 3D).
The exogenous JA treatment caused reduction in primary root length (Supplemental Fig. S1D). We observed that JA treatment induced the expression of miR166/165 significantly. To genetically confirm this, we checked the expression of miR166/165 in seedlings of coi1-16 ein2-1 pen2-4 triple mutant, which is JA insensitive30. Triple mutant roots showed reduced expression of miR166/165 in comparison to wild type (Fig. 3E).
Exogenous SA application showed inhibitory effect on primary root length (Fig. 3F). Exogenous application of SA induced the transcript level of miR166/165 in roots. To genetically confirm this, we checked its expression in npr1-2 mutant, which is insensitive to SA treatment. The endogenous SA level in npr1-2 mutant was less in comparison to wild type root31. Roots of npr1-2 show reduced expression level of miR166/165 in comparison to wild type, which further confirmed SA mediated upregulation of miR166/165 in Arabidopsis roots.
Over expression of miR166 (miR166-Oe) alters the sensitivity of roots to hormones
We have recently reported that miR166-Oe affects primary root growth through down regulation of HD-ZIP IIIs transcripts19. To investigate if the accumulation of miR166/165 and reduction in the expression of targets affected the response of roots to hormone treatment, we performed phenotypic analysis of wild type and miR166-Oe plants treated with different phytohormones (Fig. 4). The treatment with 0.1 μM IAA showed reduction in primary root length of both wild type and miR166-Oe plants in comparison to their respective untreated control, which suggest that miR166-Oe roots were less sensitive to IAA than wild type treated plants (Fig. 4A). We observed that primary root growth of miR166-Oe was more sensitive towards 1 μM ABA treatment than that of wild type plants (Fig. 4C). In order to investigate the effect of CK on root growth of miR166-Oe plants, we treated miR166-Oe line with 0.1 µM BAP and observed the phenotype. We found that primary root length of CK treated miR166-Oe plants was reduced as compared to wild type roots, suggesting that miR166-Oe plants are more sensitive to CK treatment (Fig. 4B). Additionally, primary root growth of miR166-Oe plants was more sensitive to 20 μM JA treatment (Fig. 4D). Upon treatment with 100 μM SA, less reduction in primary root length suggested that miR166-Oe plants are less sensitive to SA (Fig. 4E). Collectively these results suggest that the sensitivity of miR166-Oe plant roots vary in response to different phytohormone treatments, which could be the effect of downregulation of HD-ZIP IIIs by increased miR166/165 level in miR166-Oe plants.
Phytohormones affect the root growth of miR166-Oe and wild type plants. Primary root growth of miR166-Oe in the presence of (A) 0.1 µM IAA, (B) 0.1 µM BAP, (C) 1 µM ABA, (D) 20 µM JA, and (E) 100 µM SA. The primary root length measurement was done using Image J software from 3 dag to 7 dag. Error bars indicate ± SD (n = 10). One-way ANOVA was performed. Statistically significant differences are indicated as, * for P < 0.05,** for P < 0.01 and *** for P < 0.001.
Phytohormones differentially regulate the expression of HD-ZIP IIIs during root development
Since miR166/165 cleaves and negatively regulates HD-ZIP III transcripts, differential root growth observed in miR166-Oe plants upon phytohormone treatment might be due to the altered expression levels of HD-ZIP IIIs 19. At cellular level, the transcript level of total HD-ZIP IIIs is ultimately the result of both transcriptional regulation and post-transcriptional cleavage by miR166/165. Therefore, we analyzed the accumulation of HD–ZIP IIIs transcript level in response to various phytohormone treatments at different time intervals (1, 6, 12 and 24 hrs) by real time qRT-PCR (Fig. 5). When we checked the early response of IAA on the expression of HD–ZIP IIIs, we found an induction of expression. At 6 hrs of the IAA treatment, the expression of total HD-ZIP IIIs was only 10% of the untreated control (Fig. 5A). IAA induced the expression of total HD-ZIP IIIs at 12 and 24 hrs after treatments, in comparison to untreated control (Fig. 5A). GA treatment induces the transcript of HD-ZIP IIIs at 1 and 6 hrs, however, it reduced total HD-ZIP IIIs transcript level at 12 and 24 hrs, in comparison to untreated control (Fig. 5B). The expression of uncleaved HD-ZIP IIIs was more at 1, 12 and 24 hrs, whereas total HD-ZIP IIIs transcript level was downregulated at all the time points, except at 1 hr. The expression of HD-ZIP IIIs upon ABA treatment was induced at 1 and 6 hrs, whereas at 12 and 24 hrs the transcripts of HD-ZIP IIIs were reduced (Fig. 5C). JA treatment induces the transcript of HD-ZIP III at 1 and 12 hrs, however, transcript levels were reduced at 6 and 24 hrs of treatment (Fig. 5D).
Phytohormones affect the expression level of HD-ZIP IIIs transcript level. The expression of total HD-ZIP IIIs at 1, 6, 12 and 24 hrs of the treatment of (A) 10 µM IAA, (B) 10 µM GA, (C) 10 µM ABA, (D) 20 µM JA, (E) 100 µM SA and (F) 10 µM BAP. The control indicates untreated roots in each case. Error bars indicate ± SE of two independent biological experiments. One-way ANOVA was performed. Statistically significant differences are indicated as, * for P < 0.05,** for P < 0.01 and *** for P < 0.001. (G–M) GUS expression analysis of pPHB::PHB-GUS roots after 12 hrs of the hormone treatment in (G) untreated control (H) 10 µM IAA, (I) 10 µM GA, (J) 10 µM ABA, (K) 20 µM JA, (L) 100 µM SA, and (M) 10 µM BAP.
SA treatment induced the transcription of HD-ZIP IIIs at 1, 6 and 12 hrs, however, it reduced the expression of HD-ZIP IIIs at 24 hrs of treatment, in comparison to untreated control (Fig. 5E). Our results suggest that phytohormones regulate HD-ZIP IIIs at transcriptional level, besides post-transcriptional regulation through miR166/165 (Fig. 5N).
Expression analysis of pPHB::PHB-GUS under treatment with different phytohormones confirms hormonal regulation of HD-ZIP IIIs
Since expression level of HD-ZIP III genes was affected by different phytohormones, subsequently affecting the root development, we studied the spatial expression pattern of PHB (a member HD-ZIP IIIs) reporter pPHB::PHB-GUS in response to phytohormones. The differential expression of pPHB::PHB-GUS (a translation fusion line) in hormone treated root represents the outcome of in vivo regulation of HD-ZIP IIIs through phytohormones. This suggests that the regulation of HD-ZIP IIIs through phytohormones was stable at protein level. We analyzed GUS expression in the root meristem after 12 hrs of hormone treatment (Fig. 5G-M). It was found that GUS expression was stronger and extended upwards in the primary root tip in IAA and SA treatments, as compared to the untreated controls (Fig. 5G,H,L). GUS expression analysis confirmed the qRT-PCR results illustrating the increased expression of overall total HD-ZIP IIIs at 12 hrs upon treatment with IAA and SA. We observed reduced GUS expression after 12 hrs of BAP treatment (Fig. 5M). BAP treatment of pPHB::PHB-GUS line confirmed our qRT-PCR results, as there was a reduction in the overall total HD-ZIP IIIs expression after 12 hrs of BAP treatment (Fig. 5M). We did not observe any significant difference in GUS expression pattern upon GA, ABA and JA treatment, although differential expression pattern of total HD-ZIP IIIs transcripts was found at 12 hrs of treatment with hormones (Fig. 5I,J,K). This could be the result of differential amplification of other HD-ZIP IIIs transcripts (other than PHB) in the qRT-PCR analysis. The significant increase in pPHB::PHB-GUS expression upon SA treatment further indicates potential role of SA signaling in HD-ZIP IIIs mediated root development, in a manner opposite to IAA.
KANs show differential expression pattern in response to phytohormones
We showed that phytohormones regulate the expression of miR166/165 and its target HD-ZIP IIIs. KAN genes, members of GARP transcription factor family, regulate shoot development and organ patterning by antagonistically regulating the expression of HD-ZIP IIIs. Like leaves, the expression domain of both HD-ZIP IIIs and KANs are complementary to each other in roots20. Therefore, we also checked the effect of phytohormones on KAN genes during root growth.
We analyzed the accumulation of transcripts of KAN gene family members, KAN1, KAN2, KAN3, and KAN4, in response to hormone treatments for 1, 6, 12 and 24 hrs (Fig. 6). During IAA treatment, the expression of KAN1, KAN2, and KAN4 was found to be upregulated at 1, 6 and 12 hrs of the treatment up to 2.5 to 5 folds (Fig. 6A). At 24 hrs, the expression of all the four KANs was significantly downregulated (Fig. 6A). The expression of KAN genes did not change significantly in response to GA treatment at any time point. Among all the KAN genes, the expression of only KAN1 remained higher than the untreated control at all the time points, except at 1 hr (Fig. 6B). The expression of the KAN gene family members showed differential expression in response to ABA treatment with immediate increase at 1 and 6 hrs, followed by decrease at 12 hrs, and again increase at 24 hrs (Fig. 6C). The dynamic change in the expression of KAN genes may be due to the presence of an ABA mediated secondary regulatory mechanism. BAP treatment reduced overall expression level of total HD-ZIP IIIs at each time point, suggesting a linear temporal regulation of transcription. On the other hand, BAP treatment induced the expression level of both KANs and miR166/165 at each time point, suggesting their linear temporal regulation by CK signaling in root at transcriptional and post-transcriptional level (Figs 5F and 6D). The expression of all KAN genes was induced at 1, 6 and 12 hrs of SA treatment (Fig. 6E). However, treatment with SA for 24 hrs induced the expression of only KAN1 (Fig. 6E). JA treatment for 1 and 6 hrs induced the expression of all KANs, except KAN1 (Fig. 6F). JA treatment for 12 hrs induced the expression of all KANs, except KAN2. At 24 hrs of the treatment, the expression of all KAN genes was downregulated in comparison to wild type. Differential expression pattern of KAN genes suggests transcriptional regulation of KANs through phytohormones, which in turn would regulate transcription of HD-ZIP IIIs (Fig. 5N ).
Phytohormones affect the expression level of KAN genes. The expression of KAN genes in Arabidopsis roots was checked at 1, 6, 12 and 24 hrs of the treatment with (A) 10 µM IAA, (B) 10 µM GA, (C) 10 µM ABA, (D) 20 µM JA, (E) and 100 µM SA. The control indicates without hormone treatment in each case. Error bars indicate ± SE of two independent biological experiments. One-way ANOVA was performed. Statistically significant differences are indicated as, * for P < 0.05,** for P < 0.01 and *** for P < 0.001.
Discussion
Phytohormone signaling regulates expression of miR166/165
To study the hormone dependent regulation of miR166/165 expression, we analyzed the expression level of miR166/165 in response to different phytohormones. We found significant decrease in the expression and accumulation of mature miR166/165 as well as their precursors upon IAA treatment (Figs 1A and 2A). Auxin is a major regulatory hormone in plants and several reports have established a complex miRNA mediated crosstalk between auxin and other phytohormone signaling32. This indicates that the reduction in the expression level of miR166 in response to auxin, as evidenced by qRT-PCR and in situ localization results, is possibly affected by a complex crosstalk with other hormones (Fig. S3). Expression of miR166/165 was induced and expression of target HD-ZIP IIIs was downregulated after treatment with BAP, ABA and JA, which suggests their transcriptional regulation in root through phytohormone signaling (Figs 1 and 5). Mature miR166/165 s are derived from the precursors of their respective MIR166/165 genes, which showed differential expression pattern in response to phytohormone treatment indicating their transcriptional regulation through phytohormones.
Genetic evidences using hormone signaling mutants confirm hormonal regulation of miR166/165
Various hormone insensitive mutants showed differential expression pattern of miR166/165, which further confirmed their hormonal regulation (Fig. 3). Hormone insensitive mutants used in the present study are deficient in respective phytohormone endogenously, hence the expression patterns of miR166/165 was opposite to that of exogenous hormone treatment. In case of GA, CK, ABA, JA, and SA treatment, expression and accumulation of mature miR166/165 was found to be upregulated at 12 hrs of the treatment (Fig. 1B–F). Opposite expression of miR166/165 in hormone insensitive mutants, as shown by our qRT-PCR and whole mount in-situ hybridization experiments, further suggests their transcriptional regulation through phytohormones (Fig. 1 and Supplemental Fig. S3).
Overexpression of miR166/165 mediated reduction of HD-ZIP IIIs alters the sensitivity of roots towards phytohormones
We have previously described that primary root elongation in miR166-Oe is promoted by down regulation of HD-ZIP IIIs 19. Therefore, we analyzed the root growth of miR166-Oe in response to various phytohormones (Fig. 4). The response of miR166-Oe roots to various phytohormones indicates a correlation between levels of miR166/165 and HD-ZIP IIIs transcripts and their transcriptional regulation through phytohormones. Treatment with IAA and CK resulted in reduced root length in miR166-Oe plants, suggesting the possible involvement of miR166/165 target HD-ZIP IIIs in IAA and CK dependent root growth. In previous reports, it has been shown that HD-ZIP IIIs genes act downstream of auxin, as indicated by upregulated ATHB8 expression in shoot vasculature after auxin treatment33. Tryptophan dependent auxin biosynthesis is required for HD-ZIP IIIs expression16. This finding suggests that the effect of IAA on root growth in miR166-Oe plants is the result of altered expression of HD-ZIP IIIs genes through both transcriptional and post-transcriptional regulation. The activity of PHB and PHV is required for ISOPENTENYLTRANSFERASE 7 (IPT7) expression and CK biosynthesis and thus regulate root growth34. CK in turn represses the expression of PHB in a feedback mechanism, which was also supported by our pPHB::PHB-GUS expression result (Fig. 5M)34. Thus, the effect of CK on root growth of miR166-Oe plants could be due to the reduced activity of PHB and PHV. Treatment of miR166-Oe plants with other phytohormones like ABA, JA and SA also leads to change in root growth (Fig. 4). Expression of mature miR166/165 was upregulated upon ABA treatment and miR166-Oe roots were more sensitive to ABA treatment, suggesting possible involvement of miR166/165 in ABA mediated stress responses through negative regulation of HD-ZIP IIIs transcript level. JA and SA are known to be involved in pathogen responses in plants35. Our results have shown that the expression of miR166/165 is upregulated upon JA and SA treatment, suggesting that miR166/165 may play role in biotic and abiotic stress responses through the regulation of HD-ZIP IIIs transcript level. Our results also suggest that root growth is affected by transcriptional regulation of MIR166s and HD-ZIP IIIs genes modulated by hormones, besides post-transcriptional regulation of HD-ZIP IIIs by miR166/165.
HD-ZIP IIIs transcript level depend on post-transcriptional regulation by miR166/165 and transcriptional regulation by KANs and phytohormones
miR166/165 cleaves HD-ZIP III transcripts and differential root growth observed in miR166-Oe plants upon phytohormone treatment might be the result of altered expression levels of HD-ZIP IIIs 19. In addition to miR166/165, KAN genes also regulate the expression of HD-ZIP III genes in an antagonistic manner22. It is also likely that HD-ZIP IIIs and KANs themselves are transcriptionally regulated by multiple hormones and result in a feedback regulatory circuit with miR166/165.
We showed that the expression of total HD-ZIP IIIs remained relatively unaltered at 1 hr of auxin treatment, however, auxin treatment for 6 hrs repressed the expression. Although auxin did not induce miR166/165 expression at 6 hrs of the treatment, the expression of KANs was induced by 2.5 to 5 folds, which could downregulate HD-ZIP IIIs transcription (Fig. 5A). This indicates that an indirect regulation by KANs may contribute to the reduction in the expression of HD-ZIP IIIs genes in response to auxin treatment. The total transcripts of HD-ZIP IIIs genes increased by up to 2 folds at 12 hrs of IAA treatment (Fig. 5A), which correlates with the reduced expression of miR166/165 s (Fig. 1A). A further increase of the total transcript levels of HD-ZIP IIIs by up to 2.5 folds at 24 hrs may be largely due to the reduced expression and activity of miR166/165 and KAN genes (Fig. 5A). The expression of total HD-ZIP IIIs remained unaltered immediately after 1 hr of GA treatment, which, however, induced the expression by 2 fold at 6 hrs of treatment (Fig. 5B). Although the expression of miR166/165increased up to 2.3 fold at 6 hrs of the treatment, the upregulation of HD-ZIP IIIs genes could largely be due to their transcriptional induction upon GA treatment. The level of total HD-ZIP IIIs transcripts decreased at both 12 and 24 hrs of the GA treatment, in comparison to the untreated control (Fig. 5B). The expression of KAN genes remained relatively unaltered by GA treatment at all the three time points, suggesting no direct transcriptional regulation of KANs by GA. Additionally, ABA treatment induced the expression of HD-ZIP IIIs genes at 1 and 6 hrs of the treatments (Fig. 5C). However, ABA reduced the expression of HD-ZIP IIIs nearly equivalent to the control at 12 and 24 hrs of treatment, which could be caused by significant increase in the transcript level of miR166/165 at the same time points (Fig. 1C). ABA treatment also induced the expression of KANs showing their regulation by ABA (Fig. 6C). BAP treatment reduced the expression level of total HD-ZIP IIIs at each time points, except at 1 hr, suggesting differential temporal regulation of HD-ZIP IIIs through CK. BAP treatment induced the expression level of both KANs and miR166/165 at all the time points, suggesting their linear temporal regulation by CK signaling in root (Figs 5F and 6D). In case of JA treatment, the transcript level of total HD-ZIP IIIs was 4 fold upregulated at 1 and 12 hrs whereas, at 6 and 24 hrs the transcript level of total HD-ZIP IIIs was similar to that of wild type. The nearly similar level of total HD-ZIP IIIs transcripts in comparison to the control at 6 hrs of JA treatment correlates with the increased level of both miR166/165 and KANs (Figs 5D and 6F). However, sudden increase in the overall transcripts of HD-ZIP IIIs genes up to 4 fold at 12 hrs could be the result of their own transcriptional induction via JA (Fig. 5D). At 24 hrs of treatment, insignificant changes in total HD-ZIP IIIs transcript correlate to increased level of miR166/165 and KANs (Figs 5D and 6F). Treatment with SA showed induction of both HD-ZIP IIIs genes and miR166/165 after 1 and 6 hrs of the treatment followed by a further increase at 12 hrs (Fig. 5E). This could be due to their independent and strong transcriptional activation upon SA treatment. At 24 hrs of the treatment, nearly unchanged level of the total HD-ZIP IIIs transcripts correlates with the level of miR166/165, which might be below the control and insufficient to reduce HD-ZIP III transcripts post-transcriptionally (Fig. 5E). Thus, it seems that SA regulates the transcription of both the gene family to the similar extent. The transcript level of total HD-ZIP IIIs was induced at 6 and 12 hrs of SA treatment, however, at 24 hrs of treatment, the transcript level was significantly reduced in comparison to wild type (Fig. 5E). SA also showed strong transcriptional regulation for KANs, as evident by the induced expression level at 6 and 12 hrs of SA treatment, however, the expression of only KAN1 was found to be upregulated at 24 hrs of treatment (Fig. 6E).
In a recent report it was shown that KAN1 inhibits auxin biosynthesis, transport and signaling in a manner opposite to that of HD-ZIP IIIs 36. Gain-of-function mutant phb-1d produced shorter primary roots than wild type plants and it was shown that kan1 kan2 kan3 triple mutant displayed phenotype similar to phb-1d 36. Auxin was shown to upregulate the expression of HD-ZIP IIIs (Fig. 5G,H)37. Therefore, we hypothesize that KANs regulate root growth by auxin mediated regulation of HD-ZIP IIIs expression. We showed that the treatment with auxin led to the upregulation of HD-ZIP IIIs and KAN expression thus disturbing the KAN mediated regulation of auxin signaling, which lead to shorter roots (Supplemental Fig. S1A). Together, our results suggest that auxin dependent balanced activity of both KAN and HD-ZIP IIIs are required for proper root development. This strengthens our hypothesis that miR166/165 mediated post-transcriptional regulation of the transcript level of HD-ZIP IIIs, altered expression of KANs and a crosstalk between hormones cumulatively lead to altered root growth in response to phytohormones.
We observed a significant increase in the GUS expression of pPHB::PHB-GUS line upon SA treatment. We also showed that the level of total HD-ZIP IIIs transcripts was induced upon SA treatment. Although IAA and SA act antagonistically to each other38, we found increased expression of pPHB::PHB-GUS in both IAA and SA treatment (Fig. 5H,L). We showed that both SA and IAA induced the expression of miR166/165 (Fig. 1F). Thus, SA treatment induced the expression of both miR166/165 and its target HD-ZIP IIIs genes. Among six phytohormones analyzed in our study, IAA, CK, JA, and SA are the strongest potential transcriptional regulators of MIR166s. In contrast to other phytohormones, IAA negatively regulates expression of miR166/165.
We showed that CK treatment induced the expression level of both miR166/165 and KANs, whereas reduced the expression of total HD-ZIP IIIs. Decrease in the expression level of total HD-ZIP IIIs and induced expression of miR166/165 upon BAP treatment indicate a complex transcriptional and post-transcriptional regulation of HD-ZIP IIIs by CK. Both KANs and HD-ZIP IIIs work antagonistically by modulating the function of genes involved in auxin signaling, biosynthesis and transport, whereas the CK signaling component works in feedback loop with HD-ZIP IIIs 34. Therefore, the variation observed in the expression pattern of KANs and HD-ZIP IIIs at each time points after BAP treatment suggest their complex transcriptional regulation through a crosstalk of phytohormones signaling. JA negatively regulates root growth, which is also evident by studies with coi1-1, a JA mutant showing pleiotropic developmental defects and inhibition of root growth39.
Based on these results, we proposed a model showing complex hormonal regulation of miR166, HD-ZIP IIIs and KANs, which advocates that these genes act simultaneously during root development, involve some feedback regulation, and thus regulate root growth in Arabidopsis in a complex regulatory network (Fig. 7). Taken together, our results, in line with previous reports, suggest that miR166/165 post-transcriptionally regulate HD-ZIP IIIs, which are also regulated by various phytohormones and KANs at transcriptional level. A hormonal crosstalk along with feedback regulation by KANs and HD-ZIP IIIs, and miR166/165 maintain the balance of functional transcript level of HD-ZIP IIIs and phytohormone signaling module, which are critical for maintaining proper root growth. Increasing bodies of evidences suggests huge functional conservation and some level of diversification of miRNAs and their respective targets across plant species40. Therefore, it would be interesting to study the conservation and diversification of phytohormone mediated regulation of miR166/165, its target HD-ZIP IIIs across the plant species. Since both nutrient availability and phytohormones are known to regulate both miRNA expression and root development, it raises the possibility of regulation of these miRNAs through nutrient-phytohormone crosstalk, which remains an interesting area to be addressed1, 41.
A hypothetical model showing complex crosstalk among phytohormones, miR166/165, HD-ZIP IIIs and KAN genes regulating root growth. miR166/165 and HD-ZIP IIIs mediated root growth and development is regulated by various hormones such as IAA (red lines), GA (green lines), CK (grey lines), ABA (purple lines), JA (blue lines) and SA (orange lines) through transcriptional regulation of miR166/165, HD-ZIP IIIs and KAN genes. CK mediated regulation was adapted from34. miR166/165 post transcriptionally regulates HD-ZIP IIIs expression, whereas KANs and HD-ZIP IIIs regulate each other transcriptionally through auxin biogenesis, signaling, transport components36, 37. Solid thick lined arrows indicate strong transcriptional induction of genes. Solid thick lines with stop bars indicate strong transcriptional repression of genes. Narrower lined arrows/stop bars represent moderate or weak induction/repression, respectively. Dotted lines with arrow and repression mark represent both induction and repression of expression. Two dotted double lines (blue) with arrow/stop bars indicate hormonal regulation of root growth independent of miR166/165 and HD-ZIP IIIs.
Methods
Plant materials, growth and treatment conditions
Arabidopsis thaliana (ecotype Col-0 or Ler) and transgenic plants in either of these backgrounds were used for various experiments. Overexpression of miR166/165 (miR166-Oe) has been described earlier19. Most seed materials used in experiments were obtained from Arabidopsis Biological Resource Centre (ABRC). Growth assay was performed as described previously42. To analyze the effect of hormones on the root growth, 7 days old wild type plants were transferred onto the half strength Murashige and Skoog (MS) agar medium supplemented with phytohormones at different time point 1 hr, 6 hrs, 12 hrs and 24 hrs5, 43, 44. Mutants used in the study were axr2-1 (CS3077), gai-t6 rga-t2 rgl1-1rgl2-1rgl3-1 (CS16298), aba1-1 (CS21), arr1-3 arr12-1 (CS6981), coi1-16 ein2-1 pen2-4 (CS67818), npr1-2 (CS3801), which were first confirmed for homozygosity using PCR-based genotyping and analyzed for phenotype.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from the root tissues using TRI-reagent at different time points (Sigma-Aldrich, St. Louis, MO, USA)45. RNA isolation and qRT-PCR for the transcript level of MIR166 gene family, HD-ZIP IIIs and KAN gene family were done as described previously19, 46. Primers specific for each of the seven MIR166 (A–G) precursors were designed as described earlier47. For the analysis of transcript level of uncleaved or non-targeted HD-ZIP IIIs, primers were designed in coding regions flanking miR166/165 target sites. For the analysis of overall or total transcripts of all HD-ZIP IIIs genes, primers were designed from the conserved sites of the 5’ region of the cleavage site. The detail of primers is given in Table S1. The transcript level was normalized using ACTIN4 (ACT4) as endogenous control.
Validation of the mature miR166/165 expression profile via stem-loop qRT-PCR
The expression profile of mature miRNAs was analyzed by stem-loop qRT-PCR as described previously48. Sequence of the primers used in the reaction is given in Table S1.
Whole mount in situ hybridization
Whole mount in situ hybridization was performed with modification in the method described previously49,50,51. In brief, 7 days old seedlings were transferred to control and hormone plates and after 12 hrs, the treated seedlings were used for in situ hybridization using locked nucleic acid (LNA) probe complementary to miR166/165 (Eurogentec, Belgium). Plant tissues/samples were fixed in 1:1 mixture of paraformaldehyde based fixative (in PBS, phosphate buffer saline) and n-Heptane for 45 minutes, followed by washing twice with 100% methanol and ethanol. Tissues/samples were rehydrated by passing through gradients of ethanol (100%, 70%, 50%, and 25%) and for 10 minutes each and followed by treatments with fixative (20 minutes) and (PBS; 2 × 10 minutes). Root samples were kept for pre-hybridization for 1 hr in a hybridization mixture (50% formamide, 5X SSC, 0.1 mg/ml heparin, and 0.1% tween-20), followed by hybridization with DIG (digoxigenin)-labelled LNA probe overnight. LNA probe was labeled using DIG-Oligonucleotide 3’end leveling kit, 2nd generation (Roche Diagnostics, India), as per company’s manual. After hybridization, root samples were washed in wash buffers (containing 50% formamide, 2X SSC and 0.1% Tween-20; same without formamide, and with reduced SSC), and were incubated in anti-DIG antibody (Roche Diagnostics, India; dilution of 1:2000 times) for 4 - 12 hrs, after washing with PBT (PBS plus 0.1% Tween 20). After antibody treatment, samples were again washed with PBT, followed by incubation in detection buffer (100 mM, Tris-HCl, pH 9.5, 50 mM MgCl2, 100 mM NaCl). Detection was done by incubating with NBT/BCIP based detection solution (Roche Diagnostics, India), as per company’s manual. After visible signal was observed, the reaction was stopped with 10% glycerol, roots were mounted on slides with 10% glycerol and imaged using Nikon 80i microscope. Sequence of the miR166 LNA probe is provided in Supplemental Table S1.
References
Sarkar, A. K., Mayandi, K., Gautam, V., Barik, S. & Sarkar Das, S. Improving the Plant Root System Architecture to Combat Abiotic Stresses Incurred as a Result of Global Climate Changes. In Climate Change and Plant Abiotic Stress Tolerance 305–324 (Wiley-VCH Verlag GmbH & Co. KGaA, 2013).
Gray, W. M. Hormonal regulation of plant growth and development. PLoS Biol. 2, E311 (2004).
Saini, S., Sharma, I., Kaur, N. & Pati, P. K. Auxin: a master regulator in plant root development. Plant Cell Rep. 32, 741–57 (2013).
Overvoorde, P., Fukaki, H. & Beeckman, T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2, a001537 (2010).
Ubeda-Tomas, S. et al. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 19, 1194–9 (2009).
Ruzicka, K. et al. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA. 106, 4284–9 (2009).
Duan, L. et al. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 25, 324–41 (2013).
Raya-Gonzalez, J., Pelagio-Flores, R. & Lopez-Bucio, J. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J Plant Physiol. 169, 1348–58 (2012).
Kang, H. G. & Singh, K. B. Characterization of salicylic acid-responsive, arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J. 21, 329–39 (2000).
Depuydt, S. & Hardtke, C. S. Hormone signalling crosstalk in plant growth regulation. Curr Biol. 21, R365–73 (2011).
Meng, Y., Ma, X., Chen, D., Wu, P. & Chen, M. MicroRNA-mediated signaling involved in plant root development. Biochem Biophys Res Commun. 393, 345–9 (2010).
Han, M. H., Goud, S., Song, L. & Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 101, 1093–8 (2004).
Sakaguchi, J. & Watanabe, Y. miR165166 and the development of land plants. Dev Growth Differ. 54, 93–9 (2012).
Jung, J. H. & Park, C. M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta. 225, 1327–38 (2007).
Kim, J. et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42, 84–94 (2005).
Ursache, R. et al. Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development. 141, 1250–9 (2014).
McConnell, J. R. et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 411, 709–13 (2001).
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 465, 316–21 (2010).
Singh, A., Singh, S., Panigrahi, K. C., Reski, R. & Sarkar, A. K. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 33, 945–53 (2014).
Hawker, N. P. & Bowman, J. L. Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135, 2261–70 (2004).
Izhaki, A. & Bowman, J. L. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell. 19, 495–508 (2007).
Emery, J. F. et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 13, 1768–74 (2003).
Barik, S. et al. Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics. 103, 114–21 (2014).
Garay-Arroyo, A., De La Paz Sanchez, M., Garcia-Ponce, B., Azpeitia, E. & Alvarez-Buylla, E. R. Hormone symphony during root growth and development. Dev Dyn. 241, 1867–85 (2012).
Nagpal, P. et al. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–74 (2000).
Goh, T., Kasahara, H., Mimura, T., Kamiya, Y. & Fukaki, H. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci. 367, 1461–8 (2012).
Assmann, S. M., Snyder, J. A. & Lee, Y.-R. J. ABA-deficient (aba1) and ABA-insensitive (abi1-1, abi2-1) mutants of Arabidopsis have a wild-type stomatal response to humidity. Plant, Cell & Environment. 23, 387–395 (2000).
Mason, M. G. et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 17, 3007–18 (2005).
Argyros, R. D. et al. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 20, 2102–16 (2008).
Adams, E. & Turner, J. COI1, a jasmonate receptor, is involved in ethylene-induced inhibition of Arabidopsis root growth in the light. J Exp Bot. 61, 4373–86 (2010).
Lee, J. et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49, 79–90 (2007).
Curaba, J., Singh, M. B. & Bhalla, P. L. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot. 65, 1425–38 (2014).
Baima, S. et al. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 121, 4171–82 (1995).
Dello Ioio, R. et al. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol. 22, 1699–704 (2012).
Kunkel, B. N. & Brooks, D. M. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 5, 325–31 (2002).
Huang, T. et al. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell. 26, 246–62 (2014).
Ilegems, M. et al. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development. 137, 975–84 (2010).
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H. & Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 17, 1784–90 (2007).
Huang, Y. et al. Brassinosteroid negatively regulates jasmonate inhibition of root growth in Arabidopsis. Plant Signal Behav. 5, 140–2 (2010).
Barik, S. et al. Coevolution Pattern and Functional Conservation or Divergence of miR167s and their targets across Diverse Plant Species. Sci Rep. 5, 14611 (2015).
Rai, V. et al. Iron Availability Affects Phosphate Deficiency-Mediated Responses, and Evidence of Cross-Talk with Auxin and Zinc in Arabidopsis. Plant Cell Physiol. 56, 1107–23 (2015).
Singh, S., Singh, A., Roy, S. & Sarkar, A. K. SWP1 negatively regulates lateral root initiation and elongation in Arabidopsis. Plant Signal Behav. 7, 1522–5 (2012).
Yoon, E. K. et al. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 38, 1382–91 (2010).
Laplaze, L. et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell. 19, 3889–900 (2007).
Singh, S. et al. Sirtinol, a Sir2 protein inhibitor, affects stem cell maintenance and root development in Arabidopsis thaliana by modulating auxin-cytokinin signaling components. Sci Rep. 7, 42450 (2017).
Singh, A. et al. Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci Rep. 6, 39266 (2016).
Kim, Y. J. et al. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30, 814–22 (2011).
Gautam, V., Singh, A., Singh, S. & Sarkar, A. K. An Efficient LCM-Based Method for Tissue Specific Expression Analysis of Genes and miRNAs. Sci Rep. 6, 21577 (2016).
Hejatko, J. et al. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc. 1, 1939–46 (2006).
Sarkar, A. K. et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 446, 811–4 (2007).
Douglas, R. N. et al. ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell. 22, 1441–51 (2010).
Acknowledgements
We thank Department of Biotechnology (DBT, Govt. of India) for Ramalingaswami fellowship (BT/HRD/35/02/06/2008) grant to A.K.S. and UGC for fellowship to S.Y. Council of Scientific and Industrial Research (CSIR) provided fellowship to A.S., S.S., and V.G., and NIPGR provided internal funding, fellowship to A.K. is provided by SERB-NPDF (PDF/2015/000232) and is duly acknowledged. Fellowship to Al.S. is provided by DBT-RA and is duly acknowledged. Su.S. acknowledges NIPGR for internal grants and fellowship. SSD acknowledges Women Scientist-A (Wos-A) fellowship from Department of Science and Technology (DST), India WOS-A/LS- 1276/2014. We thank Dr. J.W. Reed, Dr. A.K. Nandi and ABRC for providing us with shy2-2, npr1-2 and other seed materials, respectively. We thank Suvakanta Barik for help with initial studies on cis-elements of MIR166s and HD-ZIP IIIs.
Author information
Authors and Affiliations
Contributions
A.S., S.R. and S.S. performed the experiments and wrote the manuscript. S.Y. performed microscopic analysis. V.G. and A.S. performed in situ hybridization. A.S., V.G., S.R., Al.S., and S.Y. performed qRT-PCR. S.S.D., Su.S. and S.R. performed growth assay experiments. A.K. performed bioinformatics analysis. A.K.S. conceptualized and designed the experiments, and revised the manuscript. All authors read and approve the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, A., Roy, S., Singh, S. et al. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana . Sci Rep 7, 3408 (2017). https://doi.org/10.1038/s41598-017-03632-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03632-w
This article is cited by
-
Overexpression of Pseudomonas putida-responsive miR393a-5p attunes morpho-physiological and biochemical parameters in Arabidopsis
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Genome-wide identification, bioinformatics and expression analysis of HD-Zip gene family in peach
BMC Plant Biology (2023)
-
miRNA mediated regulation of nitrogen response and nitrogen use efficiency of plants: the case of wheat
Physiology and Molecular Biology of Plants (2023)
-
Genome-Wide Identification of MicroRNAs that are Responsive to Virus/Viroid Infection in Nectarine Trees Through High-Throughput Sequencing
Tropical Plant Biology (2022)
-
Pi-starvation induced transcriptional changes in barley revealed by a comprehensive RNA-Seq and degradome analyses
BMC Genomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.