Abstract
High-content imaging is needed to catalog the variety of cellular phenotypes and multicellular ecosystems present in metazoan tissues. We recently developed iterative bleaching extends multiplexity (IBEX), an iterative immunolabeling and chemical bleaching method that enables multiplexed imaging (>65 parameters) in diverse tissues, including human organs relevant for international consortia efforts. IBEX is compatible with >250 commercially available antibodies and 16 unique fluorophores, and can be easily adopted to different imaging platforms using slides and nonproprietary imaging chambers. The overall protocol consists of iterative cycles of antibody labeling, imaging and chemical bleaching that can be completed at relatively low cost in 2–5 d by biologists with basic laboratory skills. To support widespread adoption, we provide extensive details on tissue processing, curated lists of validated antibodies and tissue-specific panels for multiplex imaging. Furthermore, instructions are included on how to automate the method using competitively priced instruments and reagents. Finally, we present a software solution for image alignment that can be executed by individuals without programming experience using open-source software and freeware. In summary, IBEX is a noncommercial method that can be readily implemented by academic laboratories and scaled to achieve high-content mapping of diverse tissues in support of a Human Reference Atlas or other such applications.
Similar content being viewed by others
Introduction
Ambitious efforts across multiple consortia, including the Human Cell Atlas1 and Human Biomolecular Atlas Program2, aim to characterize all cell types in the human body, with the ultimate goal of creating a Human Reference Atlas. While it is impossible to precisely estimate the number of cell types present in the human body, nor the complete list of biomarkers needed to define these unique entities, a recent report identified 1,534 anatomical structures, 622 cell types and 2,154 biomarkers (632 of which were proteins) in 11 human organs3. To provide a spatial context for these complex data, the field needs high-content methods that capture the in situ biology of diverse tissues with sufficient coverage depth. Beyond supporting efforts to build a comprehensive map of healthy human tissues, high-content imaging is critical for understanding tumor–immune interactions as well as the histopathology of disease.
To this end, several multiplexed antibody-based imaging methods have been developed4,5,6,7,8,9,10,11,12,13,14,15 and reviewed16,17,18. Furthermore, a detailed comparison of these methods can be found in a recent commentary authored by domain experts in the field of spatial proteomics19. The majority of these existing methods generate high-dimensional datasets through an iterative multistep process (a cycle) that includes: (i) immunolabeling with antibodies, (ii) image acquisition and (iii) fluorophore inactivation or antibody/chromogen removal. In addition to fluorophore-labeled antibodies, alternative methods such as immunostaining with signal amplification by exchange reaction (Immuno-SABER)13 and co-detection by indexing (CODEX)12,20,21,22 employ antibody DNA barcoding with complementary fluorescent oligonucleotides to collect multiplexed imaging data. Here, fluorescent oligonucleotides are rapidly hybridized and dehybridized to visualize multiple biomarkers in situ. In contrast to these cyclic imaging methods, technologies utilizing metal-conjugated antibodies, such as multiplexed ion beam imaging14 and imaging mass cytometry15, probe >40 markers without iterative antibody labeling or removal steps. Although commercialization of CODEX, multiplexed ion beam imaging and imaging mass cytometry have enabled deep spatial profiling of tissue samples from large clinical cohorts20,23,24, these methods are limited to specialized instruments and proprietary reagents, and, for mass spectrometry (MS)-based methods, may require highly trained engineering staff for instrument support.
For these reasons, we developed iterative bleaching extends multiplexity (IBEX)25, a high-content imaging method that can be implemented by scientists with basic laboratory skills at relatively low cost. Using commercially available reagents and microscopes, IBEX has been used to spatially characterize complex phenotypes in tissues from experimental animal models26 as well as clinically relevant human samples25. Because antibody validation and panel development can be costly in terms of time and capital19, we provide curated lists of antibodies as well as example panels for several tissues. The majority of these resources are optimized for a fixed frozen method of tissue preservation that overcomes the need for antigen retrieval; however, we demonstrate how IBEX can be adopted to formalin-fixed, paraffin-embedded (FFPE) tissues. While not documented here, we have previously shown that the basic IBEX workflow enables multiplexed imaging of heavily fixed tissues using Opal fluorophores and is compatible with commercially available oligonucleotide-conjugated antibodies25. In this paper, we provide extensive details and representative data for execution of the IBEX protocol, along with instructions on how to automate and accelerate data collection using a cost-effective fluidics device and widefield microscope. We additionally provide guidelines for the proper orientation and embedding of tissues to standardize data interpretation across specimens. To facilitate widespread adoption, we present a software solution for image alignment based on a simplified open-source interface to the Insight Toolkit (SimpleITK)27,28 that can be used by laboratory scientists without programming experience.
Development of the protocol
IBEX expands on earlier work demonstrating the feasibility of using borohydride derivatives to bleach fluorescently conjugated antibodies for multiplex imaging9,29,30. Bolognesi et al. and others have provided details on the chemistry, fluorophore inactivation and proteolytic properties of sodium borohydride under their user-defined experimental conditions9,29,30,31,32,33,34,35. For the manual IBEX method described here, we consistently found that 15 min of exposure to 1 mg/ml of lithium borohydride (LiBH4), a strong reducing agent36, eliminated fluorescence signal from the following dyes: Pacific Blue, Alexa Fluor (AF)488, AF532, phycoerythrin (PE), AF555, eFluor (eF)570, iFluor (iF)594, AF647, eF660, AF680, AF700 and AF750. Other fluorophores (e.g., fluorescein isothiocyanate) required a longer incubation time (<30 min) or bleaching in the presence of light (Brilliant Violet (BV)421 and BV510) for signal loss. Finally, certain fluorophores (Hoechst, JOJO-1, AF594 and eF615) maintained their signal over multiple bleaching and imaging cycles (Supplementary Table 1). The new automated IBEX protocol described in this report requires 0.5 mg/ml of LiBH4 treatment for 10 min or, in some cases, 20 min under constant flow (120 µl/min) for fluorophore inactivation. The continual exchange of fresh 0.5 mg/ml of LiBH4 (automated), as opposed to a one-time application of 1 mg/ml (manual), enables efficient bleaching at a reduced concentration of LiBH4 while preventing excessive bubble formation within the closed bath chamber, an issue not encountered with the manual method.
Tissue-specific panels are designed using antibodies conjugated to LiBH4-sensitive dyes with a LiBH4-resistant dye (usually Hoechst) serving as a fiducial for image alignment across iterative cycles. In our original publication, we demonstrated that LiBH4 acts by eliminating fluorescence signal by fluorophore inactivation and not antibody removal25. This feature allows for the inclusion of unconjugated primary antibodies in earlier cycles and amplification with fluorescently conjugated secondary antibodies in later cycles as outlined in the automated IBEX panels deposited here (Supplementary Table 2). While both IBEX methods are compatible with indirect immunolabeling, we prefer to use directly labeled antibodies from vendors or conjugated in house using commercial kits. In addition to the points raised by Hickey and colleagues19, we recommend purchasing >100 µg of antibody in a suitable format for conjugation, concentration of 1 mg/ml or greater in a buffer without carrier proteins or additives. For panels requiring multiple unconjugated antibodies, we recommend placing these antibodies in earlier cycles to avoid unwanted detection by the secondary antibody with directly conjugated antibodies generated in the same host. Furthermore, primary antibodies must be raised in different hosts (rabbit, goat and rat) versus multiple clones from the same host (three rabbit clones) when using unconjugated versions with secondary antibody detection. Lastly, the selected secondary antibodies should be highly cross-adsorbed against antibodies from other host species. Here, we provide additional details for pairing antibodies and fluorophores for optimal imaging using manual and automated IBEX methods (Supplementary Table 3). Importantly, the number of parameters that can be imaged per IBEX cycle is dependent on several variables, including microscope configuration, tissue autofluorescence, availability of conjugated antibodies and overall panel design (Supplementary Tables 2 and 3).
The resulting method, IBEX, provides an efficient means for evaluating the spatial landscape of diverse tissues by offering: (i) an adhesive that securely attaches tissues to the slide/coverslip surface over multiple fluid exchanges, (ii) an abbreviated antibody labeling step to shorten the overall protocol, and (iii) an open-source image registration workflow. Using the tissue adhesive chrome gelatin alum, IBEX can be performed on the most delicate of tissues without tissue loss or degradation for up to 20 cycles in some cases25. Antibody labeling is accelerated using a specialized nonheating microwave for the manual IBEX method (30–45 min per cycle) and microscope stage heater at 37 °C for the automated IBEX method (1 h per cycle). The SimpleITK workflow provides registration of images acquired as a 3D tissue stack (manual IBEX) or single 2D z-slice (automated IBEX). We document an easy-to-use Imaris XTension (Oxford Instruments) for registration that can be executed by individuals without programming experience. As an additional resource, an overview of costs is provided to aid potential users in writing an instrument grant application or adopting IBEX in their laboratory (Supplementary Table 4).
Overview of the protocol
Here, we provide detailed instructions for obtaining highly multiplexed images from a wide range of tissues using human material as exemplars (Fig. 1). The protocol outlines tissue grossing (steps 1–2, Box 1), tissue processing (steps 3–7, Box 1), manual (Box 2) or automated (Steps 8–46) IBEX imaging, and image alignment with SimpleITK (Steps 47–59). The overall workflow consists of (1) specimen preparation and preservation, (2) IBEX imaging, and (3) image processing that can be executed in 2–5 d or paused after each stage.
Specimens are first prepared using established tissue grossing protocols (steps 1–2, Box 1, 2–3 h). For the jejunum and skin samples, tissues are enlarged to show detail but do not reflect their actual size within the molds. Following sample preparation, tissues can be preserved as FFPE tissues (not shown) or as fixed frozen samples (Steps 3–7, 2 d). High-content imaging is performed using the manual (Box 2, 2–4 d) or automated (Steps 8–46, 18 h) IBEX method, consisting of iterative cycles of antibody labeling, imaging and fluorophore inactivation with LiBH4. IBEX-generated images are then processed and aligned using SimpleITK open-source software (Steps 47–59, 45 min).
Applications
The target audience for IBEX ranges from academic laboratories desiring an affordable method for highly multiplexed imaging to commercial entities seeking a scalable workflow for characterizing clinical samples. Published studies have evaluated the distribution of immune cells in the livers of prime and target vaccinated animals26 and visualized diverse stromal populations in human and mouse tissues using multiple markers25. Ongoing studies include, but are not limited to, assessing the tumor microenvironment of clinical samples and profiling unique epithelial cell populations present within the human thymus. The additional advancements described here—automation and an easy-to-use image alignment software package—make it an attractive method for gaining additional insight into the cellular ecosystems of diverse tissues.
Advantages and limitations
IBEX has several advantages over existing multiplexed antibody-based imaging methods, including: (i) an open and flexible nature, (ii) capacity to utilize different imaging platforms, (iii) short immunolabeling and bleaching time, (iv) unrestricted compatibility with hundreds of commercial antibodies conjugated to diverse fluorophores, and (v) simple implementation by biologists in standard laboratory environments. To date, we have demonstrated that 12 fluorophores, corresponding to eight distinct imaging channels, can be inactivated within 10–20 min of LiBH4 treatment alone. By comparison, methods employing H2O2 for fluorophore inactivation are limited to seven fluorophores, representing three imaging channels, and require 1 h in the presence of light for signal elimination8. Thus, the ability to inactivate diverse fluorophores in a short period of time is a substantial advantage over existing iterative imaging methods. Importantly, treatment of fixed frozen tissue sections with LiBH4 does not result in tissue loss as we previously found for H2O2 (ref. 25). In contrast to commercial systems employing DNA-barcoded antibodies (CODEX), the majority of our IBEX panels employ off-the-shelf reagents, reducing the time and cost of custom antibody conjugation37. The work outlined here and previously25 offers a resource to the field of antibody-based imaging by providing exhaustive lists of validated reagents and approved panels for mouse and human tissues. It is our intent to enable high-quality data generation while defraying the substantial cost (many thousands of US dollars for a 50-plex panel) and time (typically ~6 months) to develop equivalent panels de novo19.
The primary limitation of the original IBEX method was that it required manual execution; however, we now have an affordable automated solution for high-content imaging (Supplementary Table 4). Presently, the thickness of tissue that can be imaged is restricted to <50 µm because of local tissue warping and distortion across iterative cycles. Beyond increasing the tissue volume that can be acquired (>400 µm), future improvements include custom fabrication of imaging chambers >22 mm2 to enable automated acquisition of larger tissue areas, including whole slide scans and tissue microarrays. We are actively working on methods that increase the number of parameters that can be imaged in a single IBEX cycle by employing additional IBEX compatible fluorophores and confocal microscopes equipped with advanced detectors and extended spectral outputs of 440–790 nm. While high numerical aperture (NA) objectives (40×, 1.3) have been shown to work well in IBEX imaging, we have not yet carefully tested higher magnification (63×, 100×) or higher NA (1.4) objectives for resolving subcellular structures.
Experimental design
Tissue processing
In general, sample collection informed by anatomical landmarks (e.g., right axillary lymph node) and histological features (e.g., spatially invariant vasculature) is critical for proper specimen evaluation38. For tissue mapping efforts in support of a Human Reference Atlas, precise anatomical locations are required to integrate molecular and spatial data across individuals using a common coordinate framework2,39. For these reasons, it is important to record these details and, if possible, discuss with the contributing surgeon, radiologist and pathologist if unclear. Because the process of autolysis begins immediately after surgical removal of tissues or biopsy, we recommend placing specimens in fixative as soon as possible to prevent tissue degradation and thus preserve immunoreactivity and cytomorphology38,40. The most standard fixative, 10% neutral buffered formalin (4% formaldehyde), fixes tissue at a rate of 1–2 mm per hour at room temperature40,41. To ensure appropriate fixation, tissues should be cut into thin sections (<3–5 mm thick), placed in 15–20 fold the volume of fresh fixative, and incubated for a sufficient length of time (12–20 h) (refs. 40,41). IBEX is compatible with FFPE samples, and detailed protocols for the preparation of such samples are outlined elsewhere41. To overcome the technical challenges posed by FFPE samples, including epitope loss, the need for antigen retrieval, and high autofluorescence, we utilize a fixative with ~1% (vol/vol) formaldehyde and a gentle detergent. Following fixation, samples are immersed in 30% (wt/vol) sucrose for cryoprotection. Using this fixed frozen method, our laboratory has performed multiplexed imaging on a wide range of murine and human tissues42,43,44,45,46,47,48,49. Beyond excellent tissue preservation, this protocol significantly reduces autofluorescence by lysing red blood cells and reducing lipid-containing pigments. Prior to tissue embedding, it is important to properly orient specimens using guidelines established by surgical pathologists38,40. We provide detailed descriptions in the Procedure section for optimal handling of the human bowel, skin, lymph node and wedge resections (liver, kidney, spleen) as examples (Box 1). We are presently finalizing panels and developing workflows for a greater range of tissues, including human and mouse lung, thymus and various tumors, with our current data showing all these tissues to be compatible with the IBEX protocol.
Antibody validation and panel design
A central tenet of IBEX, and other multiplexed antibody-based imaging methods4,5,6,7,8,9,11,12,13,14,15, is the collection of reproducible imaging data using antibodies specific for their intended targets. Recently, members of multiple tissue-mapping consortia, including the Human Cell Atlas1, Human Biomolecular Atlas Program2, Human Tumor Atlas Network50, and Human Protein Atlas51, established guidelines for multiplexed antibody-based imaging methods19. We refer the reader to this work as it documents how to properly validate antibodies, build and test multiplexed antibody panels, conjugate custom antibody reagents, and overcome tissue autofluorescence. Furthermore, this work provides lists of antibody clones (>400 human; >90 mouse) that have been benchmarked by different users across distinct imaging platforms, including IBEX. An additional resource for qualifying multiplexed imaging panels can be found in a detailed protocol authored by Du and colleagues using tissue-based cyclic immunofluorescence52. Of note, this work provides several considerations for characterizing antibody immunolabeling at the pixel-, cell- and tissue-level and offers guidelines for minimizing artifacts arising from iterative imaging methods. Beyond these technical considerations, skillful panel design requires clarity on the scientific questions to be addressed, as well as knowledge of the anatomical structures and cell types present within a given organ. The Anatomical Structures, Cell Types, plus Biomarkers (ASCT+B) tables—assembled by more than 50 domain experts—are a valuable resource for understanding the tissue microanatomy of 11 human organs (bone marrow and blood, brain, heart, large intestine, kidney, lung, lymph node, skin, spleen, thymus, vasculature), with more in the pipeline3. Furthermore, one can explore these data with a state-of-the-art visualization tool, aiding in the selection of cell-specific protein biomarkers to target for multiplexed imaging (https://hubmapconsortium.github.io/ccf-asct-reporter/).
Besides these resources, we provide antibodies, multiplexed tissue panels and several IBEX-specific guidelines to aid in high-quality data generation (Supplementary Tables 1–3). Because immunolabeling can vary across tissues, fixation conditions and the imaging system employed, we recommend careful titration of all antibodies prior to IBEX imaging. Additionally, it is important to test whether novel antibodies are sensitive to LiBH4 before adding them to established multicycle panels. This can be determined by comparing immunolabeling in serial sections with or without LiBH4 pretreatment. For evaluating potential epitope loss resulting from steric hindrance, we compare the spatial distribution patterns of antibody panels acquired serially with ones acquired iteratively via IBEX, as we have previously shown25. In rare instances, if a particular antibody is thought to be impacted by LiBH4 exposure or cycle order, the affected antibody is moved to an earlier panel.
To elaborate on the effect of cycle number on antigenicity, antibodies directed against CD106 (RRID: AB_314561), Chromogranin A (RRID: AB_2892553) and DCAMKL1 (RRID: AB_873537) performed better in cycle 1 than in cycles 10, 7 and 5, respectively, in a 48-plex human thymus imaging panel (unpublished work). This issue impacts <2% of IBEX characterized antibodies versus the 12% (ref. 6) to 15–20% (ref. 8) of antibodies shown to be affected by cycle number in methods employing H2O2 as a fluorophore inactivation agent. However, we have observed diminished signal intensity with AF700-conjugated antibodies placed in later cycles (after cycle 4) as opposed to the same clone conjugated to other fluorophores. This, coupled with the limited availability of AF700-conjugated antibodies, reduces the number of parameters that can be robustly imaged over successive cycles. Given the importance of fluorophore conjugate on antibody performance, we provide guidelines for antibody–fluorophore pairing specific to manual and automated modes of IBEX (Supplementary Tables 3).
Manual IBEX
The manual IBEX method is compatible with a wide range of imaging substrates and can be easily adapted to diverse systems including upright and inverted microscopes. We have performed IBEX on upright microscopes with tissues adhered to slides. However, we prefer using an inverted microscope with a chambered coverglass, as it eliminates the need for coverslip removal between cycles. When preparing chambered coverglass samples, it is important to place the tissue in the center of the well to prevent any damage to microscope objectives. To ensure a uniform focal plane and secure adherence, the tissue must be carefully flattened onto the glass surface with a paintbrush (Extended Data Fig. 1a). For 20–30 µm tissue sections, we utilize the PELCO BioWave Pro 36500-230 microwave for antibody labeling; however, comparable immunolabeling is observed for thin (~5–10 µm) tissue sections incubated for 1 h at 37 °C. Before fluorophore inactivation with LiBH4, we extensively wash the sample (three exchanges of 1 ml of PBS) to remove mounting medium. We additionally wait until small bubbles form in the LiBH4 solution, typically 10 min after dissolving in water, before adding to the samples. Small bubbles routinely form on the tissue during the 15 min incubation of LiBH4 (Extended Data Fig. 1b). Because the manual IBEX method involves removing the sample from the microscope stage for iterative rounds of fluorophore inactivation and immunolabeling, careful attention must be paid to the alignment of images acquired over distinct cycles. We achieve this by matching shared landmarks, such as distinct nuclear morphology, in the first z-slice (Begin) and last z-slice (End) across the different image volumes (Extended Data Fig. 1c). Manual execution is required in instances where a fluidics device is not available or compatible with existing instrumentation. Indeed, the IBEX method was initially developed using an advanced confocal instrument equipped with a 405 nm laser and a white light laser source producing a continuous spectral output between 470 and 670 nm. This system confers several advantages over conventional widefield microscopes, including the ability to image more markers per cycle and the acquisition of fully resolved 3D z-stacks. However, this instrument is presently incompatible with the automated workflow described below. In addition to overcoming instrument limitations, the manual method provides users with a means to acquire highly multiplexed images under experimental conditions not amenable to IBEX automation, e.g., access to precut FFPE slides only.
Automated IBEX
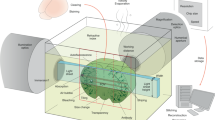
To enable widespread adoption and higher throughput, we designed an automated workflow that could be easily implemented by most laboratories and possessed an economical equipment footprint (Fig. 2a–c, Extended Data Fig. 2). We achieved this goal by integrating an inverted widefield microscope and a compact fluidics device. Our selection of an imager was based on the following technical specifications: (i) ability to image multiple fluorophores per cycle, (ii) stable multihour (>16 h) acquisitions with pixel–pixel alignment across imaging cycles, (iii) external triggering capabilities to interface with fluidics device, (iv) capture of multiple ROIs with tiling in 2D and 3D, and (v) computational clearing and adaptive deconvolution software. With regard to the fluidics device, we required an instrument that was able to send and receive transistor–transistor logic (TTL) pulses while supporting delivery of multiple solutions. The fluidics device presented here possesses ten reservoirs, and typical experiments utilize the following configuration: (i) LiBH4 in reservoir 1, (ii) Hoechst labeling in reservoir 2, (iii) antibody panels in reservoirs 3–8, and (iv) PBS wash in reservoir 10 with an empty reservoir 9. However, it is possible to deliver seven unique antibody solutions with four fluorophores per cycle to achieve a 29-parameter dataset (seven cycles of four fluorophores per cycle plus Hoechst as a fiducial) in one execution of the program. Samples are prepared using coated 22 mm square coverslips and the RC-21B closed bath imaging chamber supplied by Warner Instruments (Fig. 2b, Extended Data Fig. 2). In addition to supporting linear solution flow, the closed bath system provides a large viewing area with a total bath volume of 358 µl, greatly reducing antibody labeling costs. Together, the protocol steps of immunolabeling, imaging and fluorophore inactivation (Fig. 2c) can be automated using commercially available instruments and reagents to obtain highly multiplexed images. The ideal workflow is to start the automated IBEX protocol early in the workday. This allows for the replacement of LiBH4 and bubble removal to be completed before leaving at the end of the day, allowing the system to complete overnight.
a, The automated IBEX protocol uses a compact microfluidics system that delivers multiple solutions to an imaging chamber placed on an inverted microscope stage. Fluids are pushed through the system with an air compressor unit and delivered to the imaging chamber based on signals coordinated by the chip (2-switch). The inverted microscope and fluidics device send and receive TTL pulses using precise timings established by interfaced control software. b, Samples are sectioned onto coated coverslips and assembled into a closed bath imaging chamber. Following assembly, the imaging chamber is secured to a magnetic platform and mounted onto the microscope stage. Schematic demonstrates the multiple components of the overall sample preparation from top (1) to bottom (5). Fluid (purple shading) enters the chamber via the perfusion inlet port, collects in the bath, and exits by the perfusion outlet port that is attached to a vacuum line (not pictured). c, Automated IBEX consists of: (1) nuclear labeling with Hoechst, (2) antibody labeling for 1 h at 37 °C using a heated microscope stage, (3) imaging ROIs, (4) bleaching with LiBH4, and (5) repeating Steps 2–4 until the desired number of parameters is achieved, typically 12 h for a six-cycle, 25-plex experiment.
SimpleITK image registration
Both versions of IBEX yield a series of images that are collected separately as either 3D image stacks (manual) or 2D single z-slices (automated). Using SimpleITK, we developed an intensity-based image alignment protocol to register 2D and 3D IBEX-generated images. In our initial report, we improved upon existing algorithms53,54 by providing a workflow capable of aligning large datasets (>260 GB) that additionally offers flexibility with regard to the fiducial used for registration25. To assess the quality of image alignment, a cross correlation matrix may be generated using a marker channel that is repeated across the cycles. Here, we document a user-friendly solution for image alignment by developing an executable that is compatible with Imaris, a commercial microscopy analysis software suite in worldwide use. Through the freeware Imaris Viewer and SimpleITK Imaris XTension, biologists without programming experience can obtain cell–cell registration across x–y–z dimensions from iterative cycles of IBEX at limited cost.
Materials
Biological materials
Human tissue was obtained by following a National Institutes of Health institutional review board-approved protocol (13-C-0076) at the time of risk-reducing surgery performed as a consequence of germline genetic mutation(s). All tissue procured, which included biopsies of lymph nodes, skin, spleen, liver and jejunum, was grossly normal as determined by the operative surgeon and histopathologically normal as determined by a board-certified pathologist. Of note, all tissue was obtained within 20 min of skin incision, given our observation of neutrophil infiltrate with prolonged (>1 h) procedures. Human kidney samples were collected from patients undergoing elective renal surgery at Hannover Medical School. Samples were enrolled in this study after histologic assessment only after completion of routine diagnostics and written consent approved by the local ethics committee of Hannover Medical School (ethics-vote number 3381-16, 2893-15, 1741-13)
Caution
Any experiments involving human tissues must conform to relevant institutional and national regulations. All procedures described in this protocol were approved by an institutional review board.
Reagents
-
Tissue grossing and processing
-
BD Cytofix/Cytoperm (BD Biosciences, cat no. 554722)
-
Optimal cutting temperature (OCT) compound (Sakura, cat no. 4583)
-
Sucrose solution (see ‘Reagent setup’)
-
UltraPure agarose (Thermo Fisher Scientific, cat no. 16500-500)
-
-
Manual and automated IBEX: tissue blocking, antibody immunolabeling and fluorophore inactivation
-
Manual IBEX
-
Sample preparation (fixed frozen)
-
Chrome alum gelatin (Newcomer Supply, cat no. 1033A)
-
-
-
Automated IBEX
-
Sample preparation (Fixed frozen)
-
Chrome alum gelatin (Newcomer Supply, cat no. 1033A)
-
-
Sample preparation (FFPE)
-
AR6 buffer 10× (Akoya Biosciences, cat no. AR600250ML)
-
Chrome alum gelatin (Newcomer Supply, cat no. 1033A)
-
Ethanol, 200 proof (Decon Labs, cat no. 2701)
-
Formalin, 10% neutral buffered (Cancer Diagnostics, Inc., cat no. FX1003)
-
TBST wash buffer (see ‘Reagent setup’)
-
Xylene, histology grade (Newcomer Supply, cat no. 1446C)
-
-
Reagent setup
Sucrose solution
Dissolve 30% (wt/vol) sucrose (Millipore Sigma, cat no. S0389) in sterile 1× PBS, and pass through a 0.22 μm bottle top filter. The solution can be stored at 4 °C for several months if kept sterile.
Blocking buffer
Prepare from 1× PBS (Gibco, cat no. 10010-023) containing 0.3% Triton-X-100 (Millipore Sigma, cat no. 93443), 1% bovine serum albumin (Millipore Sigma, cat no. A1933) and 1:100 dilution of human (BD, cat no. 564219) or mouse (BD, cat no. 553141) Fc block.
TBST wash buffer
Prepare from 1× TBS (Quality Biological, cat no. 351-086-101) and 0.05% Tween 20 in diH2O (Millipore Sigma, cat no. 9005-64-5).
Equipment
-
Tissue grossing, processing and sample preparation
-
Two-well chambered coverglass (Lab-Tek, cat no. 155380)
-
Camel hair brush (Ted Pella, Inc., cat no. 11859)
-
Cell culture plate, six-well (Corning, cat no. 3335)
-
Cell Pro 500 ml 0.22 µm Bottle Top Filter (Alkali Scientific, cat no. VH50022)
-
Cryostat (Leica Biosystems, CM1950) or a comparable instrument
-
Dissecting mat, flexible, polypropylene (Newcomer Supply, cat no. 5218A)
-
Dissecting needles (Newcomer Supply, cat no. 5220PL)
-
Fine forceps (Fine Science Tools, cat no. 11412-11)
-
Forceps, custom embedding 13 cm, curved, standard grade (Newcomer Supply, cat no. 5536)
-
Histomolds, 10 × 10 × 5 mm (Sakura, cat no. 4565)
-
Histomolds, 15 × 15 × 5 mm (Sakura, cat no. 4566)
-
Low-profile microtome blades (Leica Biosystems, cat no. 14035843496)
-
Regular bevel needles 30 G (BD, cat no. 305106)
-
Scissors (Fine Science Tools, cat no. 114090-09)
-
Small digital incubator (Boekel Scientific, 133000)
-
Sterile disposable scalpels #11 (Newcomer Supply, cat no. 6802A)
-
Stereomicroscope (Zeiss, Stemi 2000-CS) or a comparable instrument
-
Stereomicroscope illuminator (Zeiss, KL 1500 LCD) or a comparable instrument
-
-
Automated IBEX
-
Sample preparation (fixed frozen)
-
Cover glass 22 × 22 mm #1.5 (Electron Microscopy Sciences, cat no. 63786-10)
-
-
Sample preparation (FFPE)
-
Cover glass 22 × 22 mm #1.5 (Electron Microscopy Sciences, cat no. 63786-10)
-
EasyDip slide staining kit (Newcomer Supply, cat no. 5300KIT)
-
EasyDip anodized aluminum jar rack holder (Newcomer Supply, cat no. 5300JRK)
-
Glass beaker, 100 ml (VWR, cat no. 10754-948)
-
Rotary microtome (Leica Biosystems, RM2255) or a comparable instrument
-
StatMark pen (Electron Microscopy Sciences, cat no. 72109-12)
-
Wash N’Dry cover slip rack (Electron Microscopy Sciences, cat no. 70366-16)
-
-
-
Manual IBEX: immunolabeling and microscopy
-
20 ml disposable syringe with Luer-lock tip (EXEL Int., cat no. 26280)
-
Millex-GS syringe filter unit (0.22 µm) (Millipore Sigma, cat no. SLGSM33SS)
-
Confocal microscope
-
The default for this protocol uses an inverted Leica TCS SP8 X confocal microscope equipped with a 40×/1.3 objective, four hybrid detectors and one photomultiplier tube detector, a white light laser source that produces a continuous spectral output between 470 and 670 nm, and an additional 405 nm laser. Images were captured at 8-bit depth, with a line average of 3, and 1,024 × 1,024 format with the following pixel dimensions: x (0.284 µm), y (0.284 µm) and z (1 µm). Images were tiled and merged using the LAS X Navigator software (LAS X 3.5.5.19976)
-
Other confocal microscopes with comparable specifications in terms of laser lines, objectives, detectors, precision stage, and control software can be used. We prefer inverted over upright microscopes because these systems eliminate the need for coverslip removal, a tedious step that can lead to tissue loss
-
-
PELCO BioWave Pro microwave (Ted Pella Inc., 36500-230) equipped with a PELCO SteadyTemp Pro (Ted Pella Inc., 50062) thermoelectric recirculating chiller
-
This specialized nonheating microwave accelerates immunolabeling without tissue loss or epitope damage
-
-
Slide moisture chamber (Scientific Device Laboratory, cat no. 197-BL)
-
-
Automated IBEX: immunolabeling and microscopy
-
31 G insulin syringes (BD, cat no. 328438)
-
20 ml disposable syringe with Luer-lock tip (EXEL Int., cat no. 26280)
-
Millex-GS syringe filter unit, 0.22 µm (Millipore Sigma, cat no. SLGSM33SS)
-
ARIA fluidics system:
-
ARIA automated perfusion system (Fluigent, cat no. CB-SY-AR-01)
-
Fluigent Low Pressure Generator (FLPG) pressure supply (Fluigent, cat no. FLPG005)
-
-
BNC Male to SMB Plug Cable RG-316 Coax in 120 inch length (two needed in total) for TTL triggering (Fairview Microwave, cat no. FMC0816315-120)
-
Imaging chamber, vacuum and heater
-
CC-28 cable assembly for heater controllers to platform (Warner Instruments, cat no. 64-0106)
-
Dedicated Workstation Vacuum System (Warner Instruments, cat no. 64-1940)
-
PM-2 Platform for Series 20 chambers, magnetic clamps, heated (Warner Instruments, cat no. 64-1561)
-
RC-21B Large Closed Bath Imaging Chamber (Warner Instruments, cat no. 64-0224)
-
SA-20PL Series 20 stage adapter (Warner Instruments, cat no. 64-0299)
-
TC-324C Single Channel Temperature Controller (Warner Instruments, cat no. 64-2400)
-
Dow Corning high-vacuum silicone grease (Millipore Sigma, cat no. Z273554)
-
10 ml size Luer-lock syringe (BD, cat no. 309604)
-
-
Leica THUNDER microscope
-
For fluorescence imaging, a custom quad-band filter with external filter wheel (PN: 11536075) with two additional single-band filters (PN: 8118215) were used to image seven dye channels per pass. The filter excitation, dichroic and emission lines are (1) Quad-Band cube: dichroic at 391/32, 479/33, 554/24, 638/31, no excitation or emission filters; (2) external filter wheel position 1: 434/32, position 2: 520/40, position 3: 585/20, position 4: 720/60, position 5: pass-through; (3) single-band 1—585/22 excitation, 594 dichroic, 625/30 emission; (4) single-band 2—635/20 excitation, 647 dichroic, 667/30 emission
-
LAS X [3.7.1.21655], the software used on the THUNDER imager, comes with the primary modules needed to perform the imaging shown here, including the THUNDER computational clearing and adaptive deconvolution routines that utilize Leica Microsystems proprietary algorithms to enhance image contrast and resolution. The additional ‘Trigger to Peripherals’ (PN: 11640613) module is required, and the ‘Dye Finder’ (PN: 11640863) module is recommended to negate spectral cross-talk. All images were captured at a 16-bit depth with the following pixel dimensions: x (0.160 µm), y (0.160 µm) and z (1 µm)
-
Other inverted widefield microscopes with comparable specifications in terms of light source and filter cubes for excitation and emission of multiple fluorophores, precision stage, adaptive focus control or other means of maintaining the same focus position over multiple cycles, objective choice, camera sensitivity and resolution, control software, deconvolution algorithms, and the ability to send and receive external triggers can be used
-
-
-
Software
-
Imaris and Imaris File Converter (x64, version 9.5.0 and higher, Bitplane)
-
Python (version 3.7.0 and higher)
-
Custom Imaris Extension (XTRegisterSameChannel, open-source software distributed under Apache 2.0 license)
-
Equipment setup
Heating stage initial install
The electrode prongs need to be carefully bent upward 30° to allow the assembled unit to fit into the stage adapter. This can be done with a pair of forceps.
Leica THUNDER microscope
The system employed in this automated IBEX protocol is a THUNDER 3D Cell Culture microscope with a high precision (Quantum) stage, a high quantum efficiency sCMOS camera (DFC9000 GTC) and a 40×/1.3 NA oil objective. The system includes an adaptive focus control unit, to ensure image focus over many hours of imaging and fluidic cycle times. The LED8 light source has eight individual LED lines for excitation with millisecond triggering. Two advanced sequencer boards were installed in the control box of the microscope configured to use SMB connections to send and receive external triggers from additional devices. A spill guard was installed on the objective turret to prevent liquids from entering the microscope body.
Automated IBEX connection of ARIA fluidics device to LAS X software
Prior to using the ARIA with LAS X, the triggers must be programmed into your hardware configuration and the cabling attached. Here we name them ‘ARIA to Leica’ and ‘Leica to ARIA’. Before imaging, set up the triggers in the hardware configuration. Within LAS X hardware configurator, navigate to the sequencer tab under ‘Configure’. Add two new triggers from the available triggers. If you do not have an open trigger on the sequencer, you will need to: (a) add a sequencer or (b) remove a triggered device. For the first trigger, give it a name (i.e., ‘Leica to ARIA’), select ‘output’ and list it as a TTL output. The second trigger will also be named (i.e., ‘ARIA to Leica’) and listed as a TTL but will be set as ‘input’. Once programmed, connect a SMB to BNC cable from the sequencer output for the ‘output’ trigger to the ‘IN’ connection on the ARIA. Connect a second SMB to BNC cable from the sequencer ‘input’ trigger to the ‘OUT’ connection of the ARIA.
Software installation for SimpleITK Imaris Python Extension
For initial setup, install Python 3.7.0 and download our Imaris extensions code repository as a zip file (https://github.com/niaid/imaris_extensions/archive/refs/heads/main.zip). Installation instructions are available online (https://github.com/niaid/imaris_extensions) and in the README.md file, which is part of the zip file. Additional details can be found in the XTRegisterSameChannel SimpleITK Imaris Python Extension YouTube tutorial (https://youtu.be/rrCajI8jroE). To illustrate the usage of this software, we provide sample data on Zenodo (https://doi.org/10.5281/zenodo.4632320).
Procedure
Manual and automated IBEX
Tissue grossing
Timing 2–3 h, depending on the number and type of samples
-
1
Survey the organ specimens for distinct anatomical landmarks that may aid in tissue orientation, and, whenever possible, preserve these metadata. If biopsies are assessed, clinical imaging data (computerized tomography scan, ultrasound) of the biopsy procedure are helpful.
-
2
Prepare samples with dissecting tools (scalpel, forceps, scissors) on a dissecting mat or other clean, fiber-free surface. For small samples, a stereomicroscope and illuminator may be required for careful dissection (Fig. 1). See Box 1 for tissue-specific details.
Critical step
Work quickly to minimize tissue damage from autolysis.
Caution
Be careful around cutting implements, and properly dispose of sharps. Use proper personal protective equipment (PPE) to reduce exposure to potential blood- or air-borne pathogens.
Tissue processing
Timing 2 d
-
3
Transfer thin (a few mm) tissue sections (skin, lymph node, wedge resections) to a 50 ml conical tube with 20 ml of fixative. Our preferred method of fixation uses 1 ml of BD Cytofix/Cytoperm for every 3 ml of 1× pH 7.4 PBS. Please consult published methods for preparing FFPE samples41. For the bowel, completely submerge the pinned tissue in fixative. The ideal ratio of fixative to tissue is 20:1. Incubate at 4 °C for 16–24 h.
Critical step
Be mindful of the expiration date published on the BD Cytofix/Cytoperm bottle. If working with potentially infectious samples, identify the fixation conditions that fully inactivate pathogen(s) before preserving samples as described above. For example, an internal biosafety review committee found that increasing the incubation period to 72 h inactivated severe acute respiratory syndrome coronavirus 2 virus particles in mouse lung tissues. Importantly, tissue epitopes were well preserved under these conditions.
Caution
Formaldehyde is a known carcinogen and environmental hazard. Wear PPE and dispose of waste properly.
-
4
Wash samples twice, <5 min per wash, in 1× PBS at room temperature (RT, 20–21 °C) to remove excess fixative.
-
5
Place sample in 30% sucrose (wt/vol), and incubate at 4 °C for 16–24 h.
-
6
Fill a Histomold with OCT medium, and embed the tissue. The tissue should occupy <60% of the mold volume. Freeze by placing the Histomold on dry ice. Using forceps, ensure the tissue is correctly oriented in the Histomold for subsequent sectioning. For orientation guidance, see Box 1.
-
7
Wrap in foil and store at −80 °C before execution of manual (Box 2) or automated IBEX protocols (Steps 8–46).
Pause point
Cryopreserved tissues can be left at −80 °C for several years.
Automated IBEX
Sample preparation
Timing 2 h for fixed frozen tissues (Step 9A); 3.5 h for FFPE tissues (Step 9B)
-
8
Coat a square 22 × 22 mm #1.5 glass coverslip by adding 3 μl of chrome alum gelatin to one side. Spread evenly using the edge of another coverslip, and dry for 30–60 min in a 60 °C oven. Denote the coated side of the coverslip using a symbol with a StatMark pen. We always mark the lower right corner with a series of characters that can only be read in one direction (QJZ). This prevents confusion if the coverslip is reoriented over the course of the experiment (sectioning, chamber assembly).
-
9
For fixed frozen tissue, follow option A; for FFPE tissue, follow option B:
-
(A)
Fixed frozen tissue preparations
-
(i)
Using a cryostat, section the OCT-embedded tissue at 10 μm thickness onto the center of the coverslip and dry overnight at RT or for 1 h at 37 °C.
Critical step
Ensure the tissue section is completely flat and centered on the glass coverslip. It is worth preparing several backup samples during one session.
Caution
Be careful around cutting implements, and properly dispose of sharps. Use PPE to reduce exposure to blood- or air-borne pathogens.
Pause point
The dried samples can be stored (covered) at RT for up to 5 d before use; however, we recommend acquiring images within 1–2 d of sample preparation.
-
(i)
-
(B)
FFPE tissue preparation
-
(i)
Use a microtome to cut sections at 5 μm.
Caution
Be careful around cutting implements, and properly dispose of sharps. Use PPE to reduce exposure to blood- or air-borne pathogens.
-
(ii)
Float tissue onto coated coverslips using the same technique utilized for FFPE sections on glass slides.
-
(iii)
Place in a 60 °C oven for 1 h to adhere the tissues to the coverslips.
-
(iv)
To deparaffinize the tissue, place coverslips in a Wash N’Dry coverslip rack. Add 100 ml of the solutions listed in (v–ix) to individual jars (eight in total) of the EasyDip slide staining kit and rack. Use forceps to transfer the coverslip rack between jars. Do not allow the tissue to dry out at any stage.
-
(v)
Incubate with 100% xylene for 10 min (100 ml in jar). Repeat this step with a fresh exchange of 100 ml 100% xylene for 10 min at RT.
-
(vi)
Incubate with 100% ethanol for 10 min (100 ml in jar) at RT.
-
(vii)
Incubate with 95% ethanol for 10 min (100 ml in jar) at RT.
-
(viii)
Incubate with 70% ethanol for 5 min with quick wash (<1 min) in water at RT.
-
(ix)
Incubate with 10% formalin for 15 min with quick wash (<1 min) in water at RT. Add Tris-buffered saline with 0.1% Tween 20 detergent (TBST), and incubate for 5 min at RT.
-
(x)
For antigen retrieval, place coverslips (in Wash N’Dry coverslip rack) in a glass beaker with 100 ml of a 1× solution of AR6 buffer (prepared in diH2O).
-
(xi)
Place beaker in a 95 °C water bath, and incubate for 40 min. Remove from the water bath, and allow to gradually cool on the bench for at least 20 min.
-
(xii)
Replace 1× AR6 buffer with 1× PBS, and cover glass beaker with aluminum foil.
Pause point
The samples can be stored in PBS at RT for up to 2 d before use.
Critical step
Always check the antigen retrieval conditions specified by the antibody vendor before immunolabeling as the buffer(s) suitable for one epitope may be incompatible with another epitope. For this reason, we optimize the antigen retrieval protocol to obtain conditions that work for all antibodies used in our experiments.
Caution
Perform deparaffinization in a properly functioning chemical hood. Read the safety data sheets associated with these chemicals, and take proper precautions before handling. Xylene and formalin (formaldehyde) are known physical and environmental hazards. Additionally, xylene and ethanol are highly flammable liquids.
-
(i)
-
(A)
Chamber assembly
Timing 15 min
-
10
Fill a 10 ml Luer-lock syringe with vacuum grease by initially removing and then replacing the plunger.
Critical step
Ensure the vacuum grease is fresh and white in color. Yellow discoloration is a sign of degradation and can lead to chamber leakage.
-
11
Coat the base of the RC-21B chamber first, by piping a line of vacuum grease 2 mm in diameter along the four sides of the square recession. Spread the grease evenly using a pipette tip.
Critical step
Avoid getting vacuum grease in the RC-21B fluid inlet or outlet ports to prevent fluid obstruction.
-
12
Apply the coverslip with adhered tissue to the base, ensuring the side with the tissue faces upward into the chamber. Gently press around the perimeter of the coverslip to ensure a continuous interface of grease surrounds, but does not spill into, the central chamber.
Critical step
If using FFPE tissue, keep the tissue wet by adding a drop of PBS onto the tissue while preparing the chamber.
-
13
Repeat Steps 11 and 12 with the following alterations: coat the top (not the base) of the RC-21B chamber with vacuum grease and use a fresh 22 × 22 mm coverslip (no tissue) to create a closed bath chamber.
-
14
Place the RC-21B chamber into the PM-2 magnetic platform with outflow side adjacent to the Warner label. Apply the magnetic clamp, keeping the Warner label on the platform base proximal to the Warner label on the magnetic clamp. See Extended Data Fig. 2.
Prepare required solutions
Timing 45 min
-
15
Add PBS directly from a 1× stock to the provided glass container in reservoir 10 of the ARIA.
-
16
Prepare Hoechst and blocking solution: add 1 μl of Hoechst 33342 and 10 μl Fc block to 1 ml of blocking buffer described in Reagents section.
-
17
Prepare antibody panels: in 400 μl total volume of blocking buffer, prepare antibody panels for each iterative cycle using previously determined titrations.
Critical step
Do not include secondary antibodies that will cross-react with primary antibodies in multiplexed panels. Always use highly cross-adsorbed secondary antibodies to prevent off-target immunolabeling.
-
18
Prepare LiBH4: prepare at least 5 ml of a 0.5 mg/ml LiBH4 solution (dissolved in diH2O).
Caution
Perform these steps in a chemical hood with appropriate PPE. The reaction can generate hydrogen gas, which is flammable. Always work with small amounts (<10 mg) of LiBH4. Replace vial of LiBH4 after 4 weeks of prolonged use as we have observed reduced bleaching efficiency after repeated exposure to air. We recommend purchasing the smallest aliquot (1 g) available. Alternative borohydride solutions may also be considered and empirically tested for their compatibility with the fluorophores and panels described here.
Critical step
Only make this solution just before the run starts. The solution becomes less effective at bleaching and will need replacement after 4 h. The solution can contain manufacturing impurities and must be passed through a 0.22 μm syringe filter before adding to the ARIA fluidics device.
Prepare the ARIA unit
Timing 10 min
-
19
Switch on the FLPG unit, and ensure a minimum of 2.2 bar is reached before switching on the ARIA unit power supply and starting the Fluigent Controller Software on the attached computer.
Critical step
If not already done, perform a calibration run and record the values, making sure these recorded values appear whenever the software is restarted.
-
20
Create a custom ARIA program based on the example steps outlined in the following table for a six-cycle experiment (total time ~630 min (10.5 h)). Load this sequence before each run.
Step
Step action
Flow rate (µl/min)
Total volume (µl)
Time (min)
TTL
Purpose
0
Prefill
3
Load the fluid lines with PBS
1
Flush tubing
200
Variable
0.5
Fill lines with PBS up to the 2-switch
2
Volume injection of PBS
120
800
~7
Rehydrate tissue
3
Volume injection of Hoechst
120
300
2.5
Nuclear labeling (Hoechst)
4
Wait
5
Nuclear labeling with Hoechst
5
Volume injection of PBS
120
600
5
PBS wash
6
Wait for user
5–10
User defines ROI and z-slice
7
Volume injection of antibody #1
120
300
2.5
Injection of antibody
8
Wait
60
Antibody immunolabeling
9
Volume injection of PBS
120
1,200
10
ARIA to THUNDER
PBS wash and signal to THUNDER for imaging
10
Wait for TTL
~10
THUNDER to ARIA
Wait for imaging to complete and signal to ARIA
11
Volume injection of PBS
120
240
2
Fluid spacer between antibodies and LiBH4
12
Volume injection of LiBH4
120
1,200
10
Fluorophore inactivation with LiBH4
13
Volume injection of PBS
120
1,200
10
PBS wash
14
Repeat steps 7–13 four times
44
Volume injection
120
300
2.5
Injection of antibody #6
45
Wait
60
Antibody immunolabeling
46
Volume injection of PBS
120
1,200
10
ARIA to THUNDER
PBS wash and signal to THUNDER for imaging
47
Wait for TTL
~10
THUNDER to ARIA
Wait for imaging to complete and signal to ARIA
48
Wait for user
Variable
Instrument paused until user input received
-
21
Add solutions to the reservoirs specified in the program on the ARIA machine. Our standard configuration is: LiBH4 (reservoir 1), Hoechst + Fc block (reservoir 2), antibody panels (reservoir 3–8) and PBS (reservoir 10).
Critical step
Only 15 ml Falcon brand tubes fit the taper to ensure an airtight seal.
Initial setup of the THUNDER microscope
Timing 5 min
-
22
Switch on microscope and LED8 source. Launch LAS X software. Select objective (this protocol is optimized for a 40×/1.30 oil immersion objective). Place the camera into 16-bit mode in the configuration tab. Select the required channels based on the fluorophores present in antibody panels. Set LED power to 25% with 150 ms exposure time for all.
Critical step
If using an AF750 filter combination, increase LED power to 40% for this channel as the signal is greatly reduced with the Adaptive Focus Control (AFC) function.
-
23
Highlight the Hoechst channel, and turn on the Linked Shading feature. Follow the command prompts using the Leica THUNDER dialog box to set this for each objective on first use.
-
24
Use the LAS X Dye Separation module to compensate for spectral spillover between channels. See Box 3 for details.
Prepare microscope stage
Timing 5 min
-
25
Insert the Warner SA-20PL stage holder into the motorized microscope stage base.
-
26
Clean the bottom coverslip of the PM-2 thoroughly with lens cleaner to remove any vacuum grease or marks. If using the recommended 40×/1.30 oil objective, coat the bottom surface of the coverslip generously with immersion oil across a wide region and avoid bubbles.
-
27
Insert and fasten the assembled imaging chamber and PM-2 magnetic platform into the stage holder. Position the assembled unit so that the Warner name is on the right side and the baseplate temperature probe hole is at the top. See Extended Data Fig. 2.
-
28
Attach the inlet tubing firmly into the RC-21B chamber.
-
29
Arrange and attach the electrical cables for the heater to the prongs on the PM-2 platform.
-
30
Insert the temperature probe into the hole at the top of the PM-2 platform.
-
31
Attach the vacuum line to the reservoir outlet, and switch on the vacuum system.
Tissue rehydration and nuclear staining
Timing 25 min
-
32
Ensure the waste line is connected to an empty container, and follow institute recommendations for waste disposal.
-
33
Prepare the LiBH4, and make a 0.5 mg/ml solution in water. Pass the dissolved solution through a 0.22 μm syringe filter. Note the time, and use within 4 h.
-
34
Switch on the electric heater, and set to 37 °C.
-
35
Launch the ARIA program.
Critical step
Following system priming, the imaging chamber will start to fill. Check there are no leaks. If air is not completely removed from the chamber during filling, loosen the stage insert and gently tilt so air bubbles rise to the outlet channel.
Final setup with triggering of the THUNDER microscope
Timing 10 min
-
36
Wait for the Fluigent software to reach the programmed pause step. By this point, the imaging chamber will be filled, the tissue will be rehydrated, and nuclear staining with Hoechst 33342 will be complete.
-
37
In the LAS X software, click the ‘Live’ button to visualize the Hoechst staining and bring the center of the tissue into sharp focus.
-
38
Click on the ‘Show Highspeed Autofocus Panel’, then choose ‘Adaptive Focus Control’ from the Focus-system dropdown options. Check ‘AFC on/off’ box so it is active. Choose ‘Continuous mode’ and AFC mode ‘Quality’.
Critical step
The green light should appear and hold position remain stable. If this fails, adjust the focus up and down. Intermittent flickering is acceptable.
-
39
Set up THUNDER Processing. Recommended parameters are to use the Instant Computational Clearing (ICC) setting with Refractive Index (RI) set to 1.33. Strength is set to 98%, and feature size is best set to 2,000 nm for membrane markers or 3,000 nm for nuclear markers in our experience.
-
40
Switch on the ‘Timecourse’ tab, and set the number of cycles to match that of the microfluidics program.
-
41
Enter Navigator mode, map the tissue using the spiral function and create the ROI for acquisition by tiling.
-
42
While in Navigator, select the triggering module.
-
43
With the ‘ARIA to Leica’ dropdown selected, check ‘Trigger linked to acquisition’, then the check box next to tile number in the Regions box. In the Channels tab, select ‘First Channel only’; in the Timelapse tab, select ‘Every cycle’; and in the Stage tab, select ‘At positions 1’.
-
44
Switch to the ‘Leica to ARIA’ dropdown, and check ‘Use in experiment’. Set number of pulses to 1 and with a 100 ms duration. Check the box next to tile number in the Regions box, then set the dropdown from Trigger to ‘After acquisition’. In the Channels tab, type the total number of channels being acquired in the ‘At Channel Number’ box. Select ‘Every Cycle’ in the Timelapse tab, and ensure the final tile number is shown under the Stage tab under ‘At positions’.
-
45
Click the Start button while in Navigator mode. The software should initialize and move to the first tile position, then pause and wait. Now click the popup box to continue the program in the Fluigent software.
Critical step
Make sure to check the ‘hold position’ button again in the AFC menu if the z-position is altered at any point.
Critical step
Bubbles form on the tissue and rise to the top of the imaging chamber during fluorophore inactivation with LiBH4, potentially impeding solution delivery to the tissue. We recommend manually removing air bubbles every four cycles. This can be done by placing a 31 G tuberculin syringe in the secondary inlet port (Fig. 2b) and aspirating while PBS is injected into the imaging chamber.
Critical step
Replace the LiBH4 after 4 h of use as its activity diminishes with time. A manual pause can be programmed into the Fluigent software.
-
46
When the cycle is complete, the file can be saved and processed by following Steps 47–59. Thoroughly clean the RC-21B chamber after each use to remove vacuum grease. Immerse in distilled water to dissolve any precipitated PBS crystals.
Image post-processing
Timing 45 min
-
47
Process raw images. We use commercially available Imaris software (Imaris x64 9.5.0) for processing raw images. Use the Imaris File Converter x64 9.5.0 software to transform the Leica confocal microscope output .lif files into .ims files.
-
48
If using the automated IBEX protocol, iterative images are included as different timepoints of the same image, e.g., timepoint 1 corresponds to cycle 1 image, timepoint 2 corresponds to cycle 2 image, and so on. To obtain individual images (antibody panel 1 from cycle 1, antibody panel 2 from cycle 2) for downstream registration, go to Edit > Crop Time, select from ‘1’ to ‘1’ for timepoint 1 and save using a name with details related to its acquisition, e.g., human_liver_panel1 or human_liver_cycle1. Open the Imaris file with all the timepoints and repeat these steps for each cycle.
-
49
For each imaging round, edit the file in Imaris and apply any desired processing steps such as image smoothing, channel naming and channel pseudo-coloring. Channel thresholding and background subtraction can be performed using the Image Processing tools in Imaris to eliminate signal from autofluorescence or left-over signal from incomplete fluorophore inactivation.
-
50
The SimpleITK registration software requires use of a consistent naming strategy. Channels should be uniformly named as prefix-separator character-postfix. Example: ‘Panel1 CD3_AF594’ (with space as a separator character for the channel where an anti-CD3 AF594 antibody was used in the first panel). See help file provided with extension and https://niaid.github.io/imaris_extensions/XTRegisterSameChannel.html for further details.
-
51
Perform image registration using Simple ITK for either manual (3D with z-stack) or automated (2D single z-slice) IBEX.
-
52
Launch the software. Click Imaris Extensions > Simple ITK > Affine Registration of z-stack using common channel.
-
53
One .ims file for each panel of the entire multiplex experiment should be uploaded using the Browse button next to File names. Enter the Channel name prefix separator character. If using the example above, this will be a single space. Click ‘Next’.
-
54
Select the registration channel. This will be the common named channel used as the fiducial marker, e.g., Hoechst. Select ‘Fixed Image’, usually the image file from cycle 1, to which other images will be registered. Select the desired directory under ‘Output file’. The ‘Start registration at resolution’ option is usually set to the maximal resolution to give the best registration possible. Reduce this if computational power is limited. Click ‘Register’ to begin.
-
55
After successful completion of the first step, the ‘Resample and Save Combined Image’ button will become available. Click and wait for the progress bar to complete.
-
56
There is a final option to generate a correlation matrix as a PDF for quantitative assessment of the registration. Check the channel box, and then click ‘Compute Correlations Before and After Registration’.
-
57
Open the output file in Imaris. The registered image of all panels with channel information should be present and is now ready for further analysis or publication.
-
58
Be patient when opening the registered file for the first time. It can take 20–30 min to view a typical six-cycle 20 GB dataset.
-
59
Immediately save the file after initial opening to speed up subsequent viewing.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Timing
-
Steps 1–2, tissue grossing: 2–3 h
-
Steps 3–7, tissue processing: 2 d
-
Box 2, manual IBEX option: total ~28.5 h
-
Steps 1–4, manual IBEX sample preparation: 2.5 h
-
Steps 5–31, manual IBEX iterative imaging: interventions and wait time (~10 h), total experimental time (26 h) for a 24-plex dataset with the following properties: 3 mm2, four cycles, 8 µm z-stack
-
-
Steps 8–46, automated IBEX option: total ~19 h
-
Steps 8–9, automated IBEX sample preparation: 2–3.5 h
-
Steps 10–21, automated IBEX chamber assembly and solution preparation: 1 h
-
Steps 22–46, automated IBEX iterative imaging: interventions and wait time (40 min), total experimental time (14 h) for a 24-plex dataset with the following properties: 3 mm2, six cycles, single z-slice
-
Steps 47–59, image post-processing: 45 min
Anticipated results
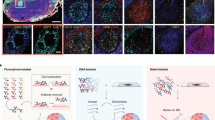
Both IBEX methods generate high-quality, multiplexed imaging data with the following attributes based on the acquisition settings described here: (i) image resolution of 0.284 (manual) or 0.160 µm (automated) in x–y, (ii) 8-bit (manual) or 16-bit (automated) dynamic range, (iii) tiled ROIs >9 mm2, and (iv) total time for an equivalent 24-plex dataset of 26 (manual, four cycles, 8 µm z-stack) or 14 h (automated, six cycles, single z-slice). These properties are distinguishing, as other imaging modalities may require 27 h to acquire a 1 mm2 area at a resolution of 0.260 µm (ref. 16). We provide here representative datasets obtained using the manual IBEX method for a wide range of human tissues, including the mesenteric lymph node (38-plex, nine cycles), spleen (25-plex, four cycles) and liver (22-plex, four cycles) (Fig. 3 and Supplementary Videos 1–3). Using the automated IBEX protocol, the following human fixed frozen tissues were profiled: mesenteric lymph node (24-plex, six cycles), jejunum (24-plex, six cycles) and skin (19-plex, five cycles), which is a very challenging tissue owing to its delicate structure and high background autofluorescence (Fig. 4a–c and Supplementary Videos 4–6). Additionally, we demonstrate an application of the automated IBEX method for imaging human kidney FFPE tissues (16-plex, five cycles) (Fig. 4d and Supplementary Video 7). Importantly, the cycle and marker numbers presented here are provided as a proof of concept and do not represent a technical limitation (see ref. 25 for IBEX imaging exceeding 65 parameters). Cell–cell alignment across imaging cycles is typical and, importantly, needed for visualizing and quantifying complex cell types in situ as phenotypic markers are frequently distributed over multiple cycles. IBEX-generated images are compatible with established methods for analyzing high-dimensional imaging data including the open-source, computational histology topography cytometry analysis toolbox (histoCAT)57, as previously demonstrated25.
a, Confocal images from a human mesenteric lymph node (LN) (nine cycles, 32 of 38 parameters shown). Scale bars: 200 µm (left), 25 µm (insets). b, Confocal images from human spleen (four cycles, 24 of 25 parameters shown). Scale bars: 200 µm (left), 25 µm (insets). Glycophorin A (Glyco A), Lumican (Lum) and Vimentin (Viment). c, Confocal images from human liver (four cycles, 11 of 22 parameters shown). Scale bars: 200 µm (left), 50 µm (insets). Glutamine synthetase (GS). See Supplementary Videos 1–3 for additional details and display of all markers.
a, Images from a human mesenteric lymph node (LN) (six cycles, 23 of 24 parameters shown). Scale bars: 200 µm (left), 50 µm (insets). b, Images from human jejunum (six cycles, 16 of 24 parameters shown). Scale bars: 200 µm (left), 50 µm (cyan box), 25 µm (red box). c, Images from human skin (five cycles, 15 of 19 parameters shown). Scale bars: 200 µm (left), 25 µm (insets). Keratin 10 (K10), Keratin 14 (K14). d, Images from FFPE kidney section (five cycles, 10 of 16 parameters shown). Scale bars: 200 µm (left), 50 µm (cyan box), 25 µm (red box). See Supplementary Videos 4–7 for additional details and display of all markers.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated during the current study are available in the Zenodo repository (https://doi.org/10.5281/zenodo.5244551).
Code availability
All custom code used in this work is freely available as open-source software under the Apache 2.0 license from the National Institute of Allergy and Infectious Diseases (NIAID) GitHub organization. The registration algorithm is available from https://github.com/niaid/sitk-ibex, and the Imaris extension code is available from https://github.com/niaid/imaris_extensions.
References
Regev, A. et al. The Human Cell Atlas. eLife https://doi.org/10.7554/eLife.27041 (2017).
Snyder, M. P. et al. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Börner, K. et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 23, 1117–1128 (2021).
Schubert, W. et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 24, 1270–1278 (2006).
Schubert, W. Topological proteomics, toponomics, MELK-technology. Adv. Biochem. Eng. Biotechnol. 83, 189–209 (2003).
Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013).
Lin, J. R., Fallahi-Sichani, M. & Sorger, P. K. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015).
Lin, J. R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife https://doi.org/10.7554/eLife.31657 (2018).
Adams, D. L., Alpaugh, R. K., Tsai, S., Tang, C. M. & Stefansson, S. Multi-Phenotypic subtyping of circulating tumor cells using sequential fluorescent quenching and restaining. Sci. Rep. 6, 33488 (2016).
Tsujikawa, T. et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 19, 203–217 (2017).
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science https://doi.org/10.1126/science.aar7042 (2018).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 e915 (2018).
Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Taube, J. M. et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J. Immunother. Cancer https://doi.org/10.1136/jitc-2019-000155 (2020).
Tan, W. C. C. et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. (Lond.) 40, 135–153 (2020).
Bodenmiller, B. Multiplexed epitope-based tissue imaging for discovery and healthcare applications. Cell Syst. 2, 225–238 (2016).
Hickey, J. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods https://doi.org/10.1038/s41592-021-01316-y (2021).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e1319 (2020).
Kennedy-Darling, J. et al. Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur. J. Immunol. 51, 1262–1277 (2021).
Phillips, D. et al. Highly multiplexed phenotyping of immunoregulatory proteins in the tumor microenvironment by CODEX tissue imaging. Front. Immunol. 12, 687673 (2021).
Keren, L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 e1319 (2018).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Radtke, A. J. et al. IBEX: a versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020).
Gola, A. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021).
Lowekamp, B. C., Chen, D. T., Ibanez, L. & Blezek, D. The design of SimpleITK. Front. Neuroinform. 7, 45 (2013).
Yaniv, Z., Lowekamp, B. C., Johnson, H. J. & Beare, R. SimpleITK Image-Analysis Notebooks: a Collaborative Environment for Education and Reproducible Research. J. Digit. Imaging 31, 290–303 (2018).
Vaughan, J. C., Jia, S. & Zhuang, X. Ultrabright photoactivatable fluorophores created by reductive caging. Nat. Methods 9, 1181–1184 (2012).
Bolognesi, M. M. et al. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J. Histochem. Cytochem. 65, 431–444 (2017).
Murray, E. et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015).
Baschong, W., Suetterlin, R. & Laeng, R. H. Control of autofluorescence of archival formaldehyde-fixed, paraffin-embedded tissue in confocal laser scanning microscopy (CLSM). J. Histochem. Cytochem. 49, 1565–1572 (2001).
Hounsell, E. F., Pickering, N. J., Stoll, M. S., Lawson, A. M. & Feizi, T. The effect of mild alkali and alkaline borohydride on the carbohydrate and peptide moieties of fetuin. Biochem. Soc. Trans. 12, 607–610 (1984).
Yakulis, V., Schmale, J., Costea, N. & Hellerp Production of Fc fragments of IgM. J. Immunol. 100, 525–529 (1968).
Corrodi, H., Hillarp, N. A. & Jonsson, G. Fluorescence methods for the histochemical demonstration of monoamines. 3. Sodium borohydride reduction of the fluorescent compounds as a specificity test. J. Histochem. Cytochem. 12, 582–586 (1964).
Nystrom, R. F., Chaikin, S. W. & Brown, W. G. Lithium borohydride as a reducing agent. J. Am. Chem. Soc. 71, 3245–3246 (1949).
Black, S. et al. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 16, 3802–3835 (2021).
Westra, W. H. Surgical Pathology Dissection: An Illustrated Guide (Springer, 2003).
Rood, J. E. et al. Toward a common coordinate framework for the human body. Cell 179, 1455–1467 (2019).
Lester, S. C. Manual of Surgical Pathology 3rd edn (Saunders, 2010).
Jonigk, D., Modde, F., Bockmeyer, C. L., Becker, J. U. & Lehmann, U. Optimized RNA extraction from non-deparaffinized, laser-microdissected material. Methods Mol. Biol. 755, 67–75 (2011).
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012).
Kastenmuller, W., Torabi-Parizi, P., Subramanian, N., Lammermann, T. & Germain, R. N. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell 150, 1235–1248 (2012).
Mao, K. et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018).
Baptista, A. P. et al. The chemoattractant receptor Ebi2 drives intranodal naive CD4+ T cell peripheralization to promote effective adaptive immunity. Immunity 50, 1188–1201 e1186 (2019).
Uderhardt, S., Martins, A. J., Tsang, J. S., Lammermann, T. & Germain, R. N. Resident macrophages cloak tissue microlesions to prevent neutrophil-driven inflammatory damage. Cell 177, 541–555 e517 (2019).
Petrovas, C. et al. Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci. Transl. Med. 9, eaag2285 (2017).
Sayin, I. et al. Spatial distribution and function of T follicular regulatory cells in human lymph nodes. J. Exp. Med. 215, 1531–1542 (2018).
Radtke, A. J. et al. Lymph-node resident CD8alpha+ dendritic cells capture antigens from migratory malaria sporozoites and induce CD8+ T cell responses. PLoS Pathog. 11, e1004637 (2015).
Srivastava, S., Ghosh, S., Kagan, J., Mazurchuk, R. & National Cancer Institute’s HTAN Implementation. The making of a PreCancer Atlas: promises, challenges, and opportunities. Trends Cancer 4, 523–536 (2018).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Du, Z. et al. Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat. Protoc. 14, 2900–2930 (2019).
Thevenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Guizar-Sicairos, M., Thurman, S. T. & Fienup, J. R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
McRae, T. D., Oleksyn, D., Miller, J. & Gao, Y. R. Robust blind spectral unmixing for fluorescence microscopy using unsupervised learning. PLoS One 14, e0225410 (2019).
Neher, R. A. et al. Blind source separation techniques for the decomposition of multiply labeled fluorescence images. Biophys. J. 96, 3791–3800 (2009).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), NIAID and National Cancer Institute (NCI). This research was also partially supported by a Research Collaboration Agreement (RCA) between NIAID and BioLegend, Inc. (RCA# 2020-0333) and the Chan Zuckerberg Initiative Human Cell Atlas Thymus Seed Network. C.J.C is supported as a UK-US Fulbright Scholar and Fight for Sight Research Scholar. Z.Y. and B.C.L. are supported by the Bioinformatics and Computational Biosciences Branch (BCBB) Support Services Contract HHSN316201300006W/HHSN27200002 to Medical Science & Computing, LLC. D.J. is supported by the grant of the European Research Council (ERC); European Consolidator Grant, XHale (reference #771883). We would like to thank R. Pelletier and M. Aruda from Fluigent for their sterling assistance with the ARIA fluidics device. We are grateful for the technical support provided by G. Portugal, E. Cox and E. Buck from Harvard Apparatus. We thank Dr. S. Pittaluga for her assistance with tissue grossing and orientation. We are appreciative of Drs. G. Cattoretti and M. Bolognesi for sharing their insights on fluorophore inactivation with sodium borohydride.
Author information
Authors and Affiliations
Contributions
A.J.R, C.J.C. and R.N.G. wrote the manuscript. A.J.R., C.J.C., H.I. and R.T.B. designed and executed the experiments. Z.R.Y. and B.L. developed image analysis software. L.Y. designed Figs. 1 and 2, and A.J.R. designed Figs. 3 and 4. A.G., A.J.R. and J.K. prepared supplementary videos. J.M. integrated the Leica microscope with the fluidics device. E.S., N.T., J.C., D.J., J.L.D. and J.M.H. provided technical insight, reagents and tissues. All authors offered guidance for the development and optimization of the workflows.
Corresponding authors
Ethics declarations
Competing interests
J.C. is an employee of Biolegend, Inc., and J.M. is an employee of Leica Microsystems, Inc.
Peer review
Peer review information
Nature Protocols thanks Fan Zhang, Yongxin Zhao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Radtke, A. J. et al. Proc. Natl Acad. Sci. USA 117, 33455–33465 (2020): https://doi.org/10.1073/pnas.2018488117
Gola, A. et al. Nature 589, 131–136 (2021): https://doi.org/10.1038/s41586-020-2977-2
Speranza, E. et al. Preprint at bioRxiv (2021): https://doi.org/10.1101/2021.09.08.459430
Extended data
Extended Data Fig. 1 Critical steps in the manual IBEX protocol.
a, Photo depicting central placement of tissue within a two-well chambered coverglass. Glass surface is coated with chrome gelatin alum (invisible when fully dry). b, Picture of small bubbles that form during successful LiBH4 treatment. c, Visual instructions on how to match unique nuclear shapes (Hoechst in yellow, blue box) across the imaging volumes. The left image corresponds to a live image in the Leica LAS X Navigator software. The right image corresponds to the image captured from the previous IBEX cycle. Red circles indicate that the described alignment procedure is being done at the first z-slice (‘Begin’) of the z-stack.
Extended Data Fig. 2 Equipment and assembly of imaging chamber for automated IBEX protocol.
a, Tissues are sectioned onto coated 22 × 22 mm square coverslips and assembled into the RC-21B Large Closed Bath Imaging Chamber. b,c, Top view of imaging chamber placed into PM-2 Platform for Series 20 chambers without (b) and with (c) magnetic platform clamp. d, Equipment footprint of automated IBEX setup; FLPG. e, Complete assembly of PM-2 Platform with RC-21B chamber onto SA-20PL Series 20 stage adapter. Fluid inlet and vacuum outlet highlight the fluid path. Heating electrodes are attached to the top and bottom of the platform using metal prongs that must be bent by user to allow placement into the stage. Temperature probe is inserted into small hole at top of platform to maintain 37 °C for the duration of the protocol.
Supplementary information
Supplementary Information
Tables 1–4 and legends for Supplementary Videos 1–7.
Supplementary Video 1
High dimensional imaging of human lymph node using manual IBEX method. Confocal images of human mesenteric lymph node from a 9 cycle 38 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 2
High dimensional imaging of human spleen using manual IBEX method. Confocal images of human spleen from a 4 cycle 25 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 3
High dimensional imaging of human liver using manual IBEX method. Confocal images of human liver from a 4 cycle 22 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 4
High dimensional imaging of human lymph node using automated IBEX method. THUNDER widefield images of human mesenteric lymph node from a 6 cycle 24 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 5
High dimensional imaging of human jejunum using automated IBEX method. THUNDER widefield images of human jejunum from a 6 cycle 24 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 6
High dimensional imaging of human skin using automated IBEX method. THUNDER widefield images of human skin from a 5 cycle 19 parameter IBEX experiment with Hoechst serving as a fiducial.
Supplementary Video 7
High dimensional imaging of human kidney (FFPE) using automated IBEX method. THUNDER widefield images of human FFPE kidney sections from a 5 cycle 16 parameter IBEX experiment with Hoechst serving as a fiducial.
Rights and permissions
About this article
Cite this article
Radtke, A.J., Chu, C.J., Yaniv, Z. et al. IBEX: an iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues. Nat Protoc 17, 378–401 (2022). https://doi.org/10.1038/s41596-021-00644-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00644-9
This article is cited by
-
Tunable PhenoCycler imaging of the murine pre-clinical tumour microenvironments
Cell & Bioscience (2024)
-
The localization of molecularly distinct microglia populations to Alzheimer's disease pathologies using QUIVER
Acta Neuropathologica Communications (2023)
-
Decoding the tumor microenvironment with spatial technologies
Nature Immunology (2023)
-
High-dimensional multi-pass flow cytometry via spectrally encoded cellular barcoding
Nature Biomedical Engineering (2023)
-
IBEX: a user-friendly and open-source solution for high-plex immunostaining
Nature Reviews Immunology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.