Abstract
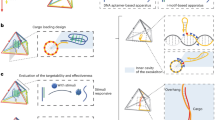
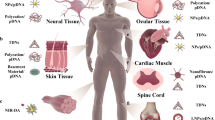
Although organic nanomaterials and inorganic nanoparticles possess inherent flexibility, facilitating functional modification, increased intracellular uptake and controllable drug release, their underlying cytotoxicity and lack of specificity still cause safety concerns. Owing to their merits, which include natural biocompatibility, structural stability, unsurpassed programmability, ease of internalization and editable functionality, tetrahedral DNA nanostructures show promising potential as an alternative vehicle for drug delivery and biomedical treatment. Here, we describe the design, fabrication, purification, characterization and potential biomedical applications of a self-assembling tetrahedral DNA nanostructure (TDN)–based multifunctional delivery system. First, relying on Watson-Crick base pairing, four single DNA strands form a simple and typical pyramid structure via one hybridization step. Then, the protocol details four different modification approaches, including replacing a short sequence of a single DNA strand by an antisense peptide nucleic acid, appending an aptamer to the vertex, direct incubation with small-molecular-weight drugs such as paclitaxel and wogonin and coating with protective agents such as cationic polymers. These modified TDN-based complexes promote the intracellular uptake and biostability of the delivered molecules, and show promise in the fields of targeted therapy, antibacterial and anticancer treatment and tissue regeneration. The entire duration of assembly and characterization depends on the cargo type and modification method, which takes from 2 h to 3 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data availability
All data generated in this study can be obtained from the corresponding author upon reasonable request.
References
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005).
Bhatia, D. et al. Icosahedral DNA nanocapsules by modular assembly. Angew. Chem. Int. Ed. Engl. 48, 4134–4137 (2009).
Winfree, E., Liu, F. R., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998).
Ranallo, S., Porchetta, A. & Ricci, F. DNA-based scaffolds for sensing applications. Anal. Chem. 91, 44–59 (2018).
Patino, T. et al. Self-sensing enzyme-powered micromotors equipped with pH-responsive DNA nanoswitches. Nano Lett. 19, 3440–3447 (2019).
Chandrasekaran, A. R. Reconfigurable DNA nanoswitches for graphical readout of molecular signals. Chem. Biochem 19, 1018–1021 (2018).
Xie, X. et al. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale 10, 5457–5465 (2018).
Madhanagopal, B. R. et al. DNA nanocarriers: programmed to deliver. Trends Biochem. Sci. 43, 997–1013 (2018).
Liang, L. et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed. Engl. 53, 7745–7750 (2014).
Li, Q. et al. Aptamer-modified tetrahedral DNA nanostructure for tumor-targeted drug delivery. ACS Appl. Mater. Interfaces 9, 36695–36701 (2017).
Lee, H. et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 7, 389–393 (2012).
Zhang, Y. et al. Inhibiting methicillin-resistant Staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 18, 5652–5659 (2018).
Walsh, A. S. et al. DNA cage delivery to mammalian cells. ACS Nano 5, 5427–5432 (2011).
Peng, Q. et al. Understanding the biomedical effects of the self-assembled tetrahedral DNA nanostructure on living cells. ACS Appl. Mater. Interfaces 8, 12733–12739 (2016).
Zhang, Q. et al. Anti-inflammatory and antioxidative effects of tetrahedral DNA nanostructures via the modulation of macrophage responses. ACS Appl. Mater. Interfaces 10, 3421–3430 (2018).
Xia, K. et al. Systematic study in mammalian cells showing no adverse response to tetrahedral DNA nanostructure. ACS Appl. Mater. Interfaces 10, 15442–15448 (2018).
Qin, X. et al. Tetrahedral framework nucleic acids prevent retina ischemia-reperfusion injury from oxidative stress via activating the Akt/Nrf2 pathway. Nanoscale 11, 20667–20675 (2019).
Liu, N. et al. Tetrahedral framework nucleic acids promote corneal epithelial wound healing in vitro and in vivo. Small 15, e1901907 (2019).
Li, J. et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 5, 8783–8789 (2011).
Shi, S. et al. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier. Nanomedicine 21, 102061 (2019).
Ma, W. et al. An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2. Nano Lett. 19, 4505–4517 (2019).
Shi, S. et al. Effects of tetrahedral framework nucleic acid/wogonin complexes on osteoarthritis. Bone Res 8, 6 (2020).
Tian, T. et al. Synthesis of an ethyleneimine/tetrahedral DNA nanostructure complex and its potential application as a multi-functional delivery vehicle. Nanoscale 9, 18402–18412 (2017).
Ge, Y. et al. PEGylated protamine-based adsorbing improves the biological properties and stability of tetrahedral framework nucleic acids. ACS Appl. Mater. Interfaces 11, 27588–27597 (2019).
Lin, M. et al. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 11, 1244–1263 (2016).
Li, J., Fan, C., Pei, H., Shi, J. & Huang, Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 25, 4386–4396 (2013).
Crommelin, D. J. A., van Hoogevest, P. & Storm, G. The role of liposomes in clinical nanomedicine development. What now? Now what? J. Control. Release 318, 256–263 (2019).
Zhang, J., Song, S., Wang, L., Pan, D. & Fan, C. A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat. Protoc. 2, 2888–2895 (2007).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Li, Y. G. et al. Controlled assembly of dendrimer-like DNA. Nat. Mater. 3, 38–42 (2004).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Lv, Y. et al. Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers. Nat. Protoc. 10, 1508–1524 (2015).
Shih, W. M., Quispe, J. D. & Joyce, G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 (2004).
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Zhang, Y. & Seeman, N. C. Construction of a DNA-truncated octahedron. J. Am. Chem. Soc. 116, 1661–1669 (1993).
Vindigni, G. et al. Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells. ACS Nano 10, 5971–5979 (2016).
Zagorovsky, K., Chou, L. Y. & Chan, W. C. Controlling DNA-nanoparticle serum interactions. Proc. Natl Acad. Sci. USA 113, 13600–13605 (2016).
Simmel, S. S., Nickels, P. C. & Liedl, T. Wireframe and tensegrity DNA nanostructures. Acc. Chem. Res 47, 1691–1699 (2014).
Shao, X. et al. Neuroprotective effect of tetrahedral DNA nanostructures in a cell model of Alzheimer’s disease. ACS Appl. Mater. Interfaces 10, 23682–23692 (2018).
Larsen, H. J., Bentin, T. & Nielsen, P. E. Antisense properties of peptide nucleic acid. Biochim. Biophys. Acta 1489, 159–166 (1999).
Haydon, D. J. et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321, 1673–1675 (2008).
RayChaudhuri, D. & Park, J. T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254 (1992).
Lin, M. et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. Engl. 54, 2151–2155 (2015).
Li, Z. et al. DNA nanostructure-based universal microarray platform for high-efficiency multiplex bioanalysis in biofluids. ACS Appl. Mater. Interfaces 6, 17944–17953 (2014).
Fu, W. et al. Enhanced efficacy of temozolomide loaded by a tetrahedral framework DNA nanoparticle in the therapy for glioblastoma. ACS Appl. Mater. Interfaces 11, 39525–39533 (2019).
Mahlknecht, G., Sela, M. & Yarden, Y. Aptamer targeting the ERBB2 receptor tyrosine kinase for applications in tumor therapy. Methods Mol. Biol. 1317, 3–15 (2015).
Yu, X. et al. Targeting EGFR/HER2/HER3 with a three-in-one aptamer-siRNA chimera confers superior activity against HER2(+) breast cancer. Mol. Ther. Nucleic Acids 10, 317–330 (2018).
Girvan, A. C. et al. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 5, 1790–1799 (2006).
Soundararajan, S. et al. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res 68, 2358–2365 (2008).
Krishna, A. G. et al. Taxol-DNA interactions: fluorescence and CD studies of DNA groove binding properties of taxol. Biochim. Biophys. Acta 1381, 104–112 (1998).
Ouameur, A. A. et al. Taxol interaction with DNA and RNA—Stability and structural features. Can. J. Chem. 82, 1112–1118 (2004).
Rusak, G. et al. Spectrophotometric analysis of flavonoid-DNA interactions and DNA damaging/protecting and cytotoxic potential of flavonoids in human peripheral blood lymphocytes. Chem. Biol. Interact. 188, 181–189 (2010).
Sun, Y. et al. Study on the interaction mechanism between DNA and the main active components in Scutellaria baicalensis Georgi. Sens. Actuators B Chem. 129, 799–810 (2008).
Khan, N. M., Ahmad, I., Ansari, M. Y. & Haqqi, T. M. Wogonin, a natural flavonoid, intercalates with genomic DNA and exhibits protective effects in IL-1β stimulated osteoarthritis chondrocytes. Chem. Biol. Interact. 274, 13–23 (2017).
Boussif, O. et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA 92, 7297–7301 (1995).
Aldrian, G. et al. PEGylation rate influences peptide-based nanoparticles mediated siRNA delivery in vitro and in vivo. J. Control. Release 256, 79–91 (2017).
Acknowledgements
This study was supported by the National Key R&D Program of China (2019YFA0110600) and the National Natural Science Foundation of China (81970916, 81671031 and 81800947).
Author information
Authors and Affiliations
Contributions
Y.L. supervised and conceived the research. T.T., W.M., Y.Z., N.L., S.S., Q.L., X.X., Q.Z., S.L., M.L. and Y.G. designed the TDN-based delivery systems and completed the corresponding experiments. T.Z., T.T., S.L., R.Z., X.C. and Y.L. interpreted data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Yonggang Ke and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Ma, W. et al. Nano Lett. 19, 4505–4517 (2019): https://doi.org/10.1021/acs.nanolett.9b01320
Zhang, Y. et al. Nano Lett. 18, 5652–5659 (2018): https://doi.org/10.1021/acs.nanolett.8b02166
Supplementary information
Supplementary Information
Supplementary Figs. 1–7
Supplementary Data 1
Source data for Fig. 3e.
Supplementary Data 2
Source data for Fig. 4f.
Supplementary Data 3
Source data for Fig. 5c.
Supplementary Data 4
Source data for Fig. 6a and b.
Supplementary Data 5
Source data for Fig. 7d and e.
Supplementary Data 6
Source data for Fig. 10a, b, d and e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, T., Tian, T., Zhou, R. et al. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat Protoc 15, 2728–2757 (2020). https://doi.org/10.1038/s41596-020-0355-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0355-z
This article is cited by
-
Tetrahedral framework nucleic acids for improving wound healing
Journal of Nanobiotechnology (2024)
-
Chemically modified microRNA delivery via DNA tetrahedral frameworks for dental pulp regeneration
Journal of Nanobiotechnology (2024)
-
Bimodal DNA self-origami material with nucleic acid function enhancement
Journal of Nanobiotechnology (2024)
-
A DNA tetrahedron-based ferroptosis-suppressing nanoparticle: superior delivery of curcumin and alleviation of diabetic osteoporosis
Bone Research (2024)
-
Tetrahedral framework nucleic acids/hyaluronic acid-methacrylic anhydride hybrid hydrogel with antimicrobial and anti-inflammatory properties for infected wound healing
International Journal of Oral Science (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.