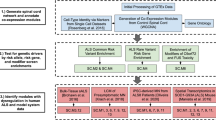
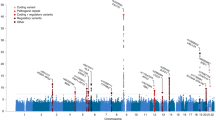
Abstract
Amyotrophic lateral sclerosis (ALS) is a progressively fatal neurodegenerative disease affecting motor neurons in the brain and spinal cord. In this study, we investigated gene expression changes in ALS via RNA sequencing in 380 postmortem samples from cervical, thoracic and lumbar spinal cord segments from 154 individuals with ALS and 49 control individuals. We observed an increase in microglia and astrocyte gene expression, accompanied by a decrease in oligodendrocyte gene expression. By creating a gene co-expression network in the ALS samples, we identified several activated microglia modules that negatively correlate with retrospective disease duration. We mapped molecular quantitative trait loci and found several potential ALS risk loci that may act through gene expression or splicing in the spinal cord and assign putative cell types for FNBP1, ACSL5, SH3RF1 and NFASC. Finally, we outline how common genetic variants associated with splicing of C9orf72 act as proxies for the well-known repeat expansion, and we use the same mechanism to suggest ATXN3 as a putative risk gene.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All raw RNA-seq data can be accessed via the National Center for Biotechnology Information’s Gene Expression Omnibus database (GSE137810, GSE124439, GSE116622 and GSE153960). Processed gene expression count matrices with de-identified metadata have been deposited on Zenodo (https://doi.org/10.5281/zenodo.6385747), and we provide an R Markdown vignette on downloading them and performing differential expression (see URLs). In addition, we provide an interactive R Shiny app to visualize the gene expression and other clinical variable associations (see URLs). Full summary statistics for expression and sQTLs have been deposited on Zenodo (https://doi.org/10.5281/zenodo.5248758). All TWAS weight files have been deposited on Zenodo (https://doi.org/10.5281/zenodo.5256613). All RNA-seq and whole-genome sequencing data generated by the NYGC ALS Consortium are made immediately available to all members of the Consortium and with other consortia with which we have a reciprocal sharing arrangement. To request immediate access to new and ongoing data generated by the NYGC ALS Consortium and for samples provided through the Target ALS Postmortem Core, complete a genetic data request form at CGND_help@nygenome.org. All whole-genome sequencing data will be deposited on dbGaP at the conclusion of the project in late 2023.
Code availability
All analysis code written in R is available in R Markdown workbooks in a GitHub repository, and specific data processing pipelines are in separate repositories (see URLs).
URLs
Website associated with this manuscript, including all code notebooks written for this project:
https://jackhump.github.io/ALS_SpinalCord_QTLs/
Gene expression counts and TPMs with de-identified metadata:
https://zenodo.org/record/6385747
Code vignette demonstrating how to download data and perform differential expression with R: https://jackhump.github.io/ALS_SpinalCord_QTLs/html/DE_Vignette.html
R Shiny app for visualization:
https://rstudio-connect.hpc.mssm.edu/als_spinal_cord_browser/
Full QTL summary statistics:
https://zenodo.org/record/5248758
Full TWAS weights:
https://doi.org/10.5281/zenodo.5256613
MSigDB:
http://www.gsea-msigdb.org/gsea/msigdb/index.jsp
Kelley et al.68 gene fidelity marker genes:
http://oldhamlab.ctec.ucsf.edu/data-download/
NeuroExpresso marker genes:
PanglaoDB marker genes:
ENCODE Blacklist:
https://github.com/Boyle-Lab/Blacklist/blob/master/lists/hg38-blacklist.v2.bed.gz
WGS QC pipeline:
https://github.com/jackhump/WGS-QC-Pipeline
QTL mapping pipeline:
https://github.com/RajLabMSSM/QTL-mapping-pipeline
DLPFC TWAS weights:
http://gusevlab.org/projects/fusion/#reference-functional-data
ExpansionHunter:
https://github.com/Illumina/ExpansionHunter
SNPnexus:
VCFs of 1000 Genomes samples: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000_genomes_project/release/20190312_biallelic_SNV_and_INDEL/
References
Ravits, J. M. & La Spada, A. R. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology 73, 805–811 (2009).
Byrne, S. et al. Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 82, 623–627 (2011).
Majounie, E. et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11, 323–330 (2012).
Renton, A. E., Chiò, A. & Traynor, B. J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 17, 17–23 (2014).
Cirulli, E. T. et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Nat. Methods 347, 1436–1441 (2016).
Kenna, K. P. et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 48, 1037–1042 (2016).
Nicolas, A. et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97, 1268–1283 (2018).
van Es, M. A. et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 41, 1083–1087 (2009).
Van Rheenen, W. et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048 (2016).
van Rheenen, W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 53, 1636–1648 (2021).
Elden, A. C. et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010).
Tazelaar, G. H. P. et al. ATXN1 repeat expansions confer risk for amyotrophic lateral sclerosis and contribute to TDP-43 mislocalization. Brain Commun. 2, fcaa064 (2020).
Lattante, S. et al. ATXN1 intermediate-length polyglutamine expansions are associated with amyotrophic lateral sclerosis. Neurobiol. Aging 64, 157.e1–157.e5 (2018).
Hirano, M. et al. Noncoding repeat expansions for ALS in Japan are associated with the ATXN8OS gene. Neurol. Genet. 4, e252 (2018).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Young, A. M. H. et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 53, 861–868 (2021).
Novikova, G. et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun. 12, 1610 (2021).
Lopes, K. et al. Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat. Genet. 54, 4–17 (2022).
Pramatarova, A., Laganière, J., Roussel, J., Brisebois, K. & Rouleau, G. A. Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J. Neurosci. 21, 3369–3374 (2001).
Jaarsma, D., Teuling, E., Haasdijk, E. D., De Zeeuw, C. I. & Hoogenraad, C. C. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J. Neurosci. 28, 2075–2088 (2008).
Yamanaka, K. et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 11, 251–253 (2008).
Lepore, A. C. et al. Focal transplantation-based astrocyte replacement is neuroprotective in a model of motor neuron disease. Nat. Neurosci. 11, 1294–1301 (2008).
Boillée, S. et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312, 1389–1392 (2006).
Wang, L., Sharma, K., Grisotti, G. & Roos, R. P. The effect of mutant SOD1 dismutase activity on non-cell autonomous degeneration in familial amyotrophic lateral sclerosis. Neurobiol. Dis. 35, 234–240 (2009).
Phatnani, H. P. et al. Intricate interplay between astrocytes and motor neurons in ALS. Proc. Natl Acad. Sci. USA 110, E756–E765 (2013).
Town, T., Nikolic, V. & Tan, J. The microglial ‘activation’ continuum: from innate to adaptive responses. J. Neuroinflammation 2, 24 (2005).
Chiu, I. M. et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Zhao, W. et al. Activated microglia initiate motor neuron injury by a nitric oxide and glutamate-mediated mechanism. J. Neuropathol. Exp. Neurol. 63, 964–977 (2004).
Haidet-Phillips, A. M. et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 29, 824–828 (2011).
Guttenplan, K. A. et al. Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nat. Commun. 11, 3753 (2020).
D’Erchia, A. M. et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 7, 10046 (2017).
Brohawn, D. G., O’Brien, L. C. & Bennett, J. P. Jr. RNAseq analyses identify tumor necrosis factor-mediated inflammation as a major abnormality in ALS spinal cord. PLoS ONE 11, e0160520 (2016).
Andrés-Benito, P., Moreno, J., Aso, E., Povedano, M. & Ferrer, I. Amyotrophic lateral sclerosis, gene deregulation in the anterior horn of the spinal cord and frontal cortex area 8: implications in frontotemporal lobar degeneration. Aging 9, 823–851 (2017).
Dols-Icardo, O. et al. Motor cortex transcriptome reveals microglial key events in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 7, e829 (2020).
Thompson, A. G. et al. Cerebrospinal fluid macrophage biomarkers in amyotrophic lateral sclerosis. Ann. Neurol. 83, 258–268 (2018).
Tanaka, H. et al. The potential of GPNMB as novel neuroprotective factor in amyotrophic lateral sclerosis. Sci. Rep. 2, 573 (2012).
Oeckl, P. et al. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol. 139, 119–134 (2020).
Hüttenrauch, M. et al. Glycoprotein NMB: a novel Alzheimer’s disease associated marker expressed in a subset of activated microglia. Acta Neuropathol. Commun. 6, 108 (2018).
Murthy, M. N. et al. Increased brain expression of GPNMB is associated with genome wide significant risk for Parkinson’s disease on chromosome 7p15.3. Neurogenetics 18, 121–133 (2017).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Halter, B. et al. Oxidative stress in skeletal muscle stimulates early expression of Rad in a mouse model of amyotrophic lateral sclerosis. Free Radic. Biol. Med. 48, 915–923 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Chen, W.-T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991 (2020).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015).
Wang, X., Park, J., Susztak, K., Zhang, N. R. & Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 380 (2019).
Hunt, G. J., Freytag, S., Bahlo, M. & Gagnon-Bartsch, J. A. dtangle: accurate and robust cell type deconvolution. Bioinformatics 35, 2093–2099 (2019).
Skene, N. G. & Grant, S. G. N. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 10, 16 (2016).
Prudencio, M. et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 18, 1175–1182 (2015).
Dickson, D. W. et al. Extensive transcriptomic study emphasizes importance of vesicular transport in C9orf72 expansion carriers. Acta Neuropathol. Commun. 7, 150 (2019).
Dolzhenko, E. et al. ExpansionHunter: a sequence-graph based tool to analyze variation in short tandem repeat regions. Preprint at https://www.biorxiv.org/content/10.1101/572545v2 (2019).
Jackson, J. L. et al. Elevated methylation levels, reduced expression levels, and frequent contractions in a clinical cohort of C9orf72 expansion carriers. Mol. Neurodegener. 15, 7 (2020).
Oldham, M. C. et al. Functional organization of the transcriptome in human brain. Nat. Neurosci. 11, 1271–1282 (2008).
Prudencio, M. et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J. Clin. Invest. 130, 6080–6092 (2020).
Klim, J. R. et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179 (2019).
Melamed, Z. et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019).
Ticozzi, N. et al. Paraoxonase gene mutations in amyotrophic lateral sclerosis. Ann. Neurol. 68, 102–107 (2010).
Li, Y. I. et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158 (2018).
de Paiva Lopes, K. et al. Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat. Genet. 54, 4–17 (2022).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Kim-Hellmuth, S. et al. Cell type-specific genetic regulation of gene expression across human tissues. Science 369, eaaz8528 (2020).
Li, Y. I., Wong, G., Humphrey, J. & Raj, T. Prioritizing Parkinson’s disease genes using population-scale transcriptomic data. Nat. Commun. 10, 994 (2019).
Kelley, K. W., Nakao-Inoue, H., Molofsky, A. V. & Oldham, M. C. Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184 (2018).
O’Rourke, J. G. et al. C9orf72 is required for proper macrophage and microglial function in mice. Science 351, 1324–1329 (2016).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Nakamura, R. et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun. Biol. 3, 526 (2020).
Paulson, H. Machado–Joseph Disease/Spinocerebellar Ataxia Type 3. in Genetic Instabilities and Neurological Diseases 2nd ed 363–377 (Academic Press, 2006).
Seidel, K. et al. Axonal inclusions in spinocerebellar ataxia type 3. Acta Neuropathol. 120, 449–460 (2010).
Prudencio, M. et al. Toward allele-specific targeting therapy and pharmacodynamic marker for spinocerebellar ataxia type 3. Sci. Transl. Med. 12, eabb7086 (2020).
Kang, S. H. et al. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat. Neurosci. 16, 571–579 (2013).
Zondler, L. et al. Peripheral monocytes are functionally altered and invade the CNS in ALS patients. Acta Neuropathol. 132, 391–411 (2016).
Saul, J. et al. Global alterations to the choroid plexus blood–CSF barrier in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 8, 92 (2020).
Månberg, A. et al. Publisher correction: Altered perivascular fibroblast activity precedes ALS disease onset. Nat. Med. 27, 1308 (2021).
Brettschneider, J. et al. Microglial activation correlates with disease progression and upper motor neuron clinical symptoms in amyotrophic lateral sclerosis. PLoS ONE 7, e39216 (2012).
Varghese, A. M. et al. Chitotriosidase, a biomarker of amyotrophic lateral sclerosis, accentuates neurodegeneration in spinal motor neurons through neuroinflammation. J. Neuroinflammation 17, 232 (2020).
Pagliardini, V. et al. Chitotriosidase and lysosomal enzymes as potential biomarkers of disease progression in amyotrophic lateral sclerosis: a survey clinic-based study. J. Neurol. Sci. 348, 245–250 (2015).
Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 51, 592–599 (2019).
Tam, O. H. et al. Postmortem cortex samples identify distinct molecular subtypes of ALS: retrotransposon activation, oxidative stress, and activated glia. Cell Rep. 29, 1164–1177 (2019).
Schroeder, A. et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3 (2006).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Bolduc, B. Quality control of reads using Trimmomatic (Cyverse) V.1. https://doi.org/10.17504/protocols.io.ewbbfan (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Harrow, J., Frankish, A., Gonzalez, J. M. & Frazer, K. A. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Li, H. et al. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Hoffman, G. E. & Schadt, E. E. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483 (2016).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Risso, D., Schwartz, K., Sherlock, G. & Dudoit, S. GC-content normalization for RNA-seq data. BMC Bioinformatics 12, 480 (2011).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012)
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019, baz046 (2019).
Mancarci, B. O. et al. Cross-laboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. eNeuro 4, ENEURO.0212-17.2017 (2017).
Mancarci, O. & French, L. Package ‘homologeneʼ: quick access to homologene and gene annotation updates. https://cran.r-project.org/web/packages/homologene/homologene.pdf (2019).
Reimand, J., Kull, M., Peterson, H., Hansen, J. & Vilo, J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007).
Love, M. I., Soneson, C. & Robinson, M. D. Importing transcript abundance datasets with tximport. https://bioconductor.statistik.tu-dortmund.de/packages/3.6/bioc/vignettes/tximport/inst/doc/tximport.html (2017).
Stegle, O., Parts, L., Piipari, M., Winn, J. & Durbin, R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 7, 500 (2012).
Taylor-Weiner, A. et al. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 20, 228 (2019).
Feng, Y.-Y. et al. RegTools: integrated analysis of genomic and transcriptomic data for discovery of splicing variants in cancer. Preprint at https://www.biorxiv.org/content/10.1101/436634v5 (2018).
Storey, J. D. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 (2003).
Oscanoa, J. et al. SNPnexus: a web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Res. 48, W185–W192 (2020).
Myers, T. A., Chanock, S. J. & Machiela, M. J. LDlinkR: an R package for rapidly calculating linkage disequilibrium statistics in diverse populations. Front. Genet. 11, 157 (2020).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2009).
Slowikowski, K. ggrepel: repulsive text and label geoms for ‘ggplot2. https://cran.microsoft.com/snapshot/2016-08-05/web/packages/ggrepel/index.html (2016).
Tang, Y., Horikoshi, M. & Li, W. ggfortify: unified interface to visualize statistical results of popular R packages. The R Journal 8, 474–485 (2016).
Pedersen, T. L. patchwork: the composer of plots. https://mran.revolutionanalytics.com/snapshot/2020-04-25/web/packages/patchwork/index.html (2019).
Xu, S. et al. Use ggbreak to effectively utilize plotting space to deal with large datasets and outliers. Front. Genet. 12, 774846 (2021).
Yin, T., Cook, D. & Lawrence, M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 13, R77 (2012).
Acknowledgements
We thank all members of the Raj laboratory for their feedback on the manuscript. This work was supported by National Institutes of Health (NIH) National Institute on Aging grants R56-AG055824 and U01-AG068880 (J.H. and T.R.), NIH National Institute of Neurological Disorders and Stroke grant U54NS123743 (J.H., T.R. and P.F.) and NIH Medical Scientist Training Program grant T3GM007280 (J.T.H.). P.F. is supported by a UK Medical Research Council Senior Clinical Fellowship and the Lady Edith Wolfson Fellowship (MR/M008606/1 and MR/S006508/1). F.K. is supported by a BOF DOCPRO fellowship from the University of Antwerp Research Fund. P.F. is supported by the UK Motor Neurone Disease Association, the Rosetrees Trust and the UCLH NIHR Biomedical Research Centre. This work was supported, in part, through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Research reported in this paper was supported by the Office of Research Infrastructure of the NIH under award numbers S10OD018522 and S10OD026880. All NYGC ALS Consortium activities are supported by the ALS Association (19-SI-459) and the Tow Foundation. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
J.H. and T.R. conceived and designed the project. J.H. led the main analysis, with S.V., R.H., J.T.H., K.P.L., F.K., K.S., M.B.B., G.N. and U.S.E. contributing code and performing additional data analyses. J.H. and T.R. oversaw all aspects of the study, with input from D.A.K., H.P. and P.F. D.F. and H.P. designed the sample collection methodology, reviewed sample and data quality and coordinated NYGC ALS Consortium postmortem RNA research activity. The NYGC ALS Consortium and the Target ALS Human Postmortem Tissue Core provided human tissue samples as well as pathological, genetic and clinical information. J.H. wrote the manuscript, with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Sali Farhan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Results, Discussion and Methods and Supplementary Figs. 1–26
Supplementary Table
Supplementary Tables 1–15 and Supplementary Acknowledgements
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Humphrey, J., Venkatesh, S., Hasan, R. et al. Integrative transcriptomic analysis of the amyotrophic lateral sclerosis spinal cord implicates glial activation and suggests new risk genes. Nat Neurosci 26, 150–162 (2023). https://doi.org/10.1038/s41593-022-01205-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01205-3