Abstract
The hippocampal CA2 region is essential for social memory. To determine whether CA2 activity encodes social interactions, we recorded extracellularly from CA2 pyramidal neurons (PNs) in male mice during social behavior. Although CA2 neuronal firing showed only weak spatial selectivity, it accurately encoded contextual changes and distinguished between a novel and a familiar mouse. In the Df(16)A+/− mouse model of the human 22q11.2 microdeletion, which confers a 30-fold increased risk of schizophrenia, CA2 social coding was impaired, consistent with the social memory deficit observed in these mice; in contrast, spatial coding accuracy was greatly enhanced. CA2 PNs were previously found to be hyperpolarized in Df(16)A+/− mice, likely due to upregulation of TREK-1 K+ current. We found that TREK-1 blockade rescued social memory and CA2 social coding in Df(16)A+/− mice, supporting a crucial role for CA2 in the normal encoding of social stimuli and in social behavioral dysfunction in disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data and code availability
The datasets generated and/or analyzed in the current study are available from the corresponding author upon reasonable request.
All scripts for analyzing data are also available upon reasonable request.
References
Berry, R. J. & Bronson, F. H. Life history and bioeconomy of the house mouse. Biol. Rev. 67, 519–550 (1992).
Meltzer, H. Y., Thompson, P. A., Lee, M. A. & Ranjan, R. Neuropsychologic deficits in schizophrenia: relation to social function and effect of antipsychotic drug treatment. Neuropsychopharmacology 14, 27S–33S (1996).
Steinvorth, S., Levine, B. & Corkin, S. Medial temporal lobe structures are needed to re-experience remote autobiographical memories: evidence from H. M. and W. R. Neuropsychologia 43, 479–496 (2005).
Kogan, J. H., Frankland, P. W. & Silva, A. J. Long-term memory underlying hippocampus- dependent social recognition in mice. Hippocampus 1063, 47–56 (2000).
Squire, L. R. & Wixted, J. T. The cognitive neuroscience of human memory since H. M. Annu. Rev. Neurosci. 34, 259–288 (2011).
O’Keefe, J. & Dostrovsky, J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175 (1971).
MacDonald, C. J., Lepage, K. Q., Eden, U. T. & Eichenbaum, H. Hippocampal ‘time cells’ bridge the gap in memory for discontiguous events. Neuron 71, 737–749 (2011).
Kraus, B., Robinson, R., White, J., Eichenbaum, H. & Hasselmo, M. Hippocampal ‘time cells’: time versus path integration. Neuron 78, 1090–1101 (2013).
McKenzie, S. et al. Hippocampal representation of related and opposing memories develop within distinct, hierarchically organized neural schemas. Neuron 83, 202–215 (2014).
Meira, T. et al. A hippocampal circuit linking dorsal CA2 to ventral CA1 critical for social memory dynamics. Nat. Commun. 9, 1–14 (2018).
Hitti, F. L. & Siegelbaum, S. A. The hippocampal CA2 region is essential for social memory. Nature 508, 88–92 (2014).
Stevenson, E. L. & Caldwell, H. K. Lesions to the CA2 region of the hippocampus impair social memory in mice. Eur. J. Neurosci. 40, 3294–3301 (2014).
Okuyama, T., Kitamura, T., Roy, D. S., Itohara, S. & Tonegawa, S. Ventral CA1 neurons store social memory. Science 353, 1536–1541 (2016).
Rao, R. P., von Heimendahl, M., Bahr, V. & Brecht, M. Neuronal responses to conspecifics in the ventral CA1. Cell Rep. 27, 3460–3472 (2019).
Mankin, E. A., Diehl, G. W., Sparks, F. T., Leutgeb, S. & Leutgeb, J. K. Hippocampal CA2 activity patterns change over time to a larger extent than between spatial contexts. Neuron 85, 190–202 (2015).
Alexander, G. M. et al. Social and novel contexts modify hippocampal CA2 representations of space. Nat. Commun. 7, 10300 (2016).
Oliva, A., Fern, A. & Ber, A. Spatial coding and physiological properties of hippocampal neurons in the Cornu Ammonis subregions. Hippocampus 1607, 1593–1607 (2016).
Lu, L. et al. Topography of place maps along the CA3-to-CA2 axis of the hippocampus. Neuron 87, 1078–1092 (2015).
Wintzer, M. E., Boehringer, R., Polygalov, D. & McHugh, T. J. The hippocampal CA2 ensemble is sensitive to contextual change. J. Neurosci. 34, 3056–3066 (2014).
Benes, F. M., Kwok, E. W., Vincent, S. L. & Todtenkopf, M. S. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biol. Psychiatry 44, 88–97 (1998).
Zhang, Z. J. & Reynolds, G. P. A selective decrease in the relative density of parvalbumin- immunoreactive neurons in the hippocampus in schizophrenia. Schizophr. Res. 55, 1–10 (2002).
Piskorowski, R. A. et al. Age-dependent specific changes in area CA2 of the hippocampus and social memory deficit in a mouse model of the 22q11.2 deletion syndrome. Neuron 89, 163–176 (2016).
Karayiorgou, M., Simon, T. J. & Gogos, J. A. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 11, 402–416 (2010).
Talley, E. M., Solo, G., Lei, Q., Kim, D. & Bayliss, D. A. CNS distribution of members of the two-pore-domain (KCNK) potassium channel family. J. Neurosci. 21, 7491–7505 (2001).
Boehringer, R. et al. Chronic loss of CA2 transmission leads to hippocampal hyperexcitability. Neuron 94, 642–655 (2017).
Skaggs, W. E. & McNaughton, B. L. Spatial firing properties of hippocampal CA1 populations in an environment containing two visually identical regions. J. Neurosci. 18, 8455–8466 (1998).
Spiers, H. J., Hayman, R. M. A., Jovalekic, A., Marozzi, E. & Jeffery, K. J. Place field repetition and purely local remapping in a multicompartment environment. Cereb. Cortex 25, 10–25 (2015).
Grieves, R. M., Duvelle, É., Wood, E. R. & Dudchenko, P. A. Field repetition and local mapping in the hippocampus and the medial entorhinal cortex. J. Neurophysiol. 118, 2378–2388 (2017).
Stefanini, F. et al. A distributed neural code in the dentate gyrus and in CA1. Neuron 107, 703–716 (2020).
Keinath, A. T. et al. Precise spatial coding is preserved along the longitudinal hippocampal axis. Hippocampus 24, 1533–1548 (2014).
Saitta, L. Support-vector networks. Mach. Learn. 297, 273–297 (1995).
Barak, O., Rigotti, M. & Fusi, S. The sparseness of mixed selectivity neurons controls the generalization–discrimination trade-off. J. Neurosci. 33, 3844–3856 (2013).
Rigotti, M. et al. The importance of mixed selectivity in complex cognitive tasks. Nature 497, 585–590 (2013).
Giocomo, L. M. et al. Grid cells use HCN1 channels for spatial scaling. Cell 147, 1159–1170 (2011).
Borsotto, M. et al. Targeting two-pore domain K+ channels TREK-1 and TASK-3 for the treatment of depression: a new therapeutic concept. Br. J. Pharmacol. 172, 771–784 (2015).
Voloshyna, I. et al. TREK-1 Is a novel molecular target in prostate cancer. Cancer Res. 68, 1197–1204 (2008).
Di Pasquale, G. et al. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 9, 1306–1312 (2003).
Smith, A. S., Williams Avram, S. K., Cymerblit-Sabba, A., Song, J. & Young, W. S. Targeted activation of the hippocampal CA2 area strongly enhances social memory. Mol. Psychiatry 21, 1137–1144 (2016).
von Heimendahl, M., Rao, R. P. & Brecht, M. Weak and nondiscriminative responses to conspecifics in the rat hippocampus. J. Neurosci. 32, 2129–2141 (2012).
Stark, K. L. et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 40, 751–760 (2008).
Chiang, M. C., Huang, A. J. Y., Wintzer, M. E., Ohshima, T. & McHugh, T. J. A role for CA3 in social recognition memory. Behav. Brain Res. 354, 22–30 (2018).
Talbot, Z. N. et al. Normal CA1 place fields but discoordinated network discharge in a Fmr1-null mouse model of fragile X syndrome. Neuron 97, 684–697 (2018).
Zaremba, J. D. et al. Impaired hippocampal place cell dynamics in a mouse model of the 22q11.2 deletion. Nat. Neurosci. 20, 1612 (2017).
Raam, T., Mcavoy, K. M., Besnard, A., Veenema, A. H. & Sahay, A. Hippocampal oxytocin receptors are necessary for discrimination of social stimuli. Nat. Commun. 8, 2001 (2017).
Young, W. S., Li, J., Wersinger, S. R. & Palkovits, M. The vasopressin 1b receptor is prominent in the hippocampal area CA2 where it is unaffected by restraint stress or adrenalectomy. Neuroscience 143, 1031–1039 (2006).
Cui, Z., Gerfen, C. R. & Young, W. S. Hypothalamic and other connections with dorsal CA2 area of the mouse hippocampus. J. Comp. Neurol. 521, 1844–1866 (2013).
Kay, K. et al. A hippocampal network for spatial coding during immobility. Nature 531, 185–190 (2016).
Alexander, G. M. et al. CA2 neuronal activity controls hippocampal low gamma and ripple oscillations. eLife 7, e38052 (2018).
Schmitzer-torbert, N. & Redish, A. D. Neuronal activity in the rodent dorsal striatum in sequential navigation: separation of spatial and reward responses on the multiple T task. J. Neurophysiol. 91, 2259–2272 (2004).
Brown, E. N., Frank, L. M., Tang, D., Quirk, M. C. & Wilson, M. A. A statistical paradigm for neural spike train decoding applied to position prediction from ensemble firing patterns of rat hippocampal place cells. J. Neurosci. 18, 7411–7425 (1998).
Acknowledgements
We thank J. Gogos for initially providing the Df(16)A+/− mice and for advice and guidance. We thank Y. Matsushita of Ono Pharmaceuticals for suggesting that we study the action of spadin. We also thank Y.M. Zafrina, A. Kleinbort and H.G. Yueh for their technical support, B. Santoro for assistance with animal breeding and C.D. Salzman and D. Aranov for helpful discussions and comments on the manuscript. This work was supported by a grant from the National Science Foundationʼs Graduate Research Fellowship Program (to M.L.D.), by grants R01MH104602 and R01MH106629 (to S.A.S.), a grant DBI-1707398 from the NeuroNex program of NSF (to S.F.), a grant from the Zegar Family Foundation (to S.A.S.) and a grant from Ono Pharmaceuticals (to S.A.S.).
Author information
Authors and Affiliations
Contributions
M.L.D., J.A.G., S.F. and S.A.S. designed the experiments and analyses. M.L.D. performed the in vivo recordings, and M.L.D. and T.M. performed the behavioral experiments. M.L.D. and F.S. analyzed the data. M.L.D. and S.A.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Caswell Barry and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Single CA2 cell spatial firing properties during the different sessions of the three- chamber task.
a–d, Single cell measures that did not differ during sessions (ANOVA p > 0.05; n = 192 CA2 neurons from 6 mice). e, Burst index (number of spikes in bursts of at least three successive spikes with an interspike interval < 6 ms) was significantly higher in sessions with novel objects and the novel mouse (p = 0.002, 0.001 two-sided t-test with Bonferroni correction post hoc to performing ANOVA; n = 192 CA2 neurons from 6 mice). Box plots display the center line as the mean; box limits are upper and lower quartiles; whiskers show min to max values in data sets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
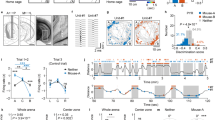
Extended Data Fig. 2 Mouse movement behavior and CA2 and CA1 neuron spatial decoding properties in various conditions.
a–b, There was no significant difference between (a) mean velocity (ANOVA, p = 0.36, n = 6,3,5,5 mice), or (b) average path length (ANOVA, p = 0.71, n = 6,3,5,5 mice) in the different experimental groups of mice. Data is presented as mean ± SEM. c, The relationship between the number of cells used to decode position by an SVM linear classifier and the accuracy of decoding for CA1 (left) and CA2 (right) cells. Note: CA2 spatial decoding accuracy was not significantly greater than chance levels in any of our recordings for any number of cells. CA1 decoding accuracy became greater than chance as additional cells were added to the decoder. Even with 30 CA2 cells, spatial decoding accuracy was less than chance and less than decoding accuracy with half as many CA1 cells. d, Example plots of real (black) versus predicted position by the model (red). e, CA1 population activity decoded position significantly better than chance in all sessions of the 3-chamber task. Data shown for two mice (CA1_1, n = 21 neurons; CA1_2, n = 27 neurons). f, CA2 population activity did not decode position better than chance in any 3-chamber task session (left graphs) or during the four individual ten-minute sessions of the pre-habituation session (right graphs). Data shown for two mice (CA2_2, n = 25 neurons; CA2_3, n = 31 neurons). Box plots display the center line as the mean; box limits are upper and lower quartiles; whiskers show min to max values in data sets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Extended Data Fig. 3 Bayesian Decoding of Position in the three-chamber task.
a, Bayesian decoding of position based on CA1 activity (dark-shaded bars) in one example animal was significantly greater than chance performance (light-shaded bars) in all five sessions of the three-chamber task. Example shown for one wild-type animal. (*p < 0.05, **p < 0.01, **p < 0.001, two-sided Wilcoxon Rank Sum). b, Example Bayesian decoding of position from CA2 activity in two animals in the three-chamber task. For all 6 wild-type animals examined, the Bayesian decoder for position based on CA2 neuron activity never performed significantly better than chance. (p > 0.05, two-sided Wilcoxon Rank Sum). P values relative to chance for all WT mice with CA2 recordings were equal to: WT7, p = 0.48; WT2, p = 0.36; WT3, p = 0.06; WT5, p = 0.17; WT10, p = 0.19; WT9, p = 0.49. Box plots display the center line as the mean; box limits are upper and lower quartiles; whiskers show min to max values in data sets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Extended Data Fig. 4 The firing rate of a subset of CA2 cells significantly increased from the non-social to the social sessions.
a, An example cell that only began to fire in the social sessions. b, Firing rate distributions for all CA2 cells in nonsocial (blue bars) and social (orange bars) sessions, each fit with a Gaussian distribution. Six percent of CA2 cells that were active in the social sessions were classified as silent in the non-social sessions based on firing rates >2 SD below the median firing rate (<0.007 Hz). c, Mean CA2 firing rates in the social versus non-social sessions in the three-chamber task. Each circle is a different cell. Red circles, the 40 cells whose z-scored mean firing rates increased >2-fold in social versus non-social sessions. Blue circles, the 3 cells whose z-scored firing rates decreased >2-fold in the social versus non-social sessions. There was a significant increase in mean firing rate (two-sided paired t-test, p = 0.015) from the non-social (mean firing rate=1.69 Hz, sem=0.19) to the social sessions (mean firing rate=1.84 Hz, sem=0.19). d, Change in z-scored firing rate from the empty arena session to the social sessions (right) and to the novel object session (left). The two firing rate vectors differed significantly (two-sided Wilcoxon rank-sum test, p = 5.80 e-37). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
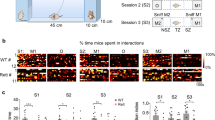
Extended Data Fig. 5 Responses of CA1 and CA2 neurons to novel stimuli.
a, Color-coded z-scored firing rates in the interaction zone around the novel mouse (session 4) and familiar mouse (sessions 3 and 5) for 87 CA1 neurons (n = 3 mice). There was no significant difference in firing rates among sessions (ANOVA, p = 0.17). b, Cumulative distributions of z-scored firing rates for all 192 CA2 neurons (n = 6 mice) in the interaction zones in each session of the five 3-chamber task sessions. CA2 activity in the interaction zones was significantly different among the different sessions (Kruskal Wallis test; p < 0.0001). CA2 firing rates in interaction zone around the novel object (wire cup cage) were significantly greater than in same spatial location in empty chamber (two-sided Mann-Whitney test, p = 0.02). CA2 firing rates around the novel mouse were significantly higher than around the novel object (mean z-scored firing rates=0.83 ± .06 and 0.19 ± 0.08, respectively; two-sided Mann-Whitney test, p = 0.56).
Extended Data Fig. 6 CA2 spatial firing in Df(16)A+/− mice and effect of spadin.
a, Additional examples of CA2 neuron spatial firing from wild-type, Df(16)A+/−, and spadin-treated Df(16)A+/− animals. Each row under a given group shows pairs of heatmap and trajectory plots in the five sessions for an individual neuron from a given animal identified on the left. Maximum firing rate (Hz) is indicated above the heatmaps; numbers of cells from each animal in each group are indicated in the tables above. b-f, CA2 neuron firing and spatial properties. Red bars, wild-type mice; blue bars, Df(16)A+/− mice; orange bars, Df(16)A+/− mice injected with spadin. b, Df(16)A+/− mice have a lower overall firing rate than wild-type mice (p = 0.02, paired t-test). Spadin increased the firing rate in the Df(16)A+/− mice (p = 0.04, two-sided paired t-test). c-f, Compared to CA2 neurons in wild-type mice, CA2 neurons in Df(16)A+/− mice had: c, fewer place fields per cell (p < 0.0001, paired t-test); d, smaller place fields (p = 0.02); e, place fields with higher spatial selectivity (p = 0.02); and f, place fields with higher spatial information content (p = 0.008). CA2 neuron firing rates, place field size, selectivity and spatial information content in Df(16)A+/− mice treated with spadin were not significantly different from values in wild-type mice (p > 0.05). Number of fields per cell in spadin-treated Df(16)A+/− mice was significantly greater than in untreated Df(16)A+/− animals (p = 0.01) but significantly less than in wild-type mice (p = 0.02). For all statistics listed: WT, n = 192 neurons; Df(16)A+/−, n = 128 neurons; Df(16)A+/− given spadin, n = 91 neurons. Box plots display the center line as the mean; box limits are upper and lower quartiles; whiskers show min to max values in data sets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Extended Data Fig. 7 Control experiments for firing properties of CA2 neurons in Df(16)A+/− mice.
a,b, Saline injection (control for spadin injection) in Df(16)A+/− mice did not alter CA2 (a) mean firing rate (two-sided paired t-test p = 0.21) or (b) spatial stability in the five sessions of the three-chamber task (two-sided paired t-test, p = 0.31; n = 128 neurons). c, CA2 neuron firing rate in saline-injected Df(16)A+/− mice did not differ in interaction zone around the novel compared to familiar mouse (two-sided paired t-test, p = 0.7), similar to uninjected Df(16)A+/− mice (Fig. 7a) but distinct from CA2 novel firing preference in wild-type mice (Fig. 4c) and spadin-injected Df(16)A+/− mice (Fig. 7d). d–f, CA2 neuron firing properties in two groups of wild-type control mice used for comparison with Df(16)A+/− mice (all on identical C57Bl/6 J backgrounds): wild-type littermates of Df(16)A+/− mice (n = 56 neurons from 2 mice) and wild-type non-littermates (n = 136 neurons from 4 mice). d, There was no significant difference in mean firing rate between wild-type non-littermates (WT) and wild-type littermates (LM) (paired t-test, p = 0.19). e, CA2 spatial stability was slightly but significantly lower in wild-type littermates compared to wild-type non-littermates (two-sided paired t-test with Bonferroni correction, p = 0.002); spatial stability of both wild-type groups was significantly less than that of Df(16)A+/− mice (two-sided paired t-test with Bonferroni correction, p < 0.0001 in both cases). f, The two wild-type control groups did not differ in their increase in firing around the novel compared to familiar animal (two-sided paired t-test, p = 0.45). Box plots display the center line as the mean; box limits are upper and lower quartiles; whiskers show min to max values in data sets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
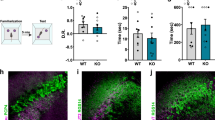
Extended Data Fig. 8 Effects of TREK-1 inhibition on social behavior.
a, Control experiment for direct interaction test using two different novel mice in trials 1 and 2. b, There was no decrease in exploration of the second novel mouse when spadin was administered 30 min before trial 1 (p = 0.57, two-sided paired t-test, n = 6 mice). c, There was no decrease in exploration of the second novel mouse in trial 2 in Df(16)A+/− mice expressing TREK-1 DN in CA2 (p = 0.34, two-sided paired t-test, n = 6 mice). Black points and lines show individual animals. Red lines and points show means. Bars show SEM. d–f, Comparison of social interaction times in three-chamber task (d) for wild-type mice, Df(16)A+/− mice, and Df(16)A+/− mice injected with spadin (n = 6, 5 and 5 mice, respectively). e, Time spent in interaction zone around cup containing the novel animal (novel session 4; bars with dark shades of color) compared to time spent in same interaction zone when the familiar mouse was present (averaged from familiar sessions 3 and 5; bars with light shades of color). Wild-type and spadin-treated Df(16)A+/− groups spent significantly less time exploring the familiar animal (two-sided paired t-test, p < 0.01). Df(16)A+/− mice spent similar time exploring the familiar and novel animals (p > 0.05; paired t-test). f, Data from panel e plotted as difference scores. (WT versus Df(16)A+/− mice, p = 0.007, df=3,F = 1.2; Df(16)A+/− mice in absence versus presence of spadin, p = 0.03, post hoc to ANOVA, p = 0.02, df=3,F = 1.2). Box plots display the center line as the mean; box limits are upper and lower quartiles; whiskers show min to max values in data sets. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Extended Data Fig. 9 Tetrode tracks from all animals in which recordings were obtained for this study.
Targeting of CA2 and CA1 in the animals included in this study. Some of our 8 tetrodes targeted to CA2 may have picked up a few cells from neighboring CA1 and CA3 regions, which would only cause us to underestimate any true differences between CA2 and CA1 spatial and social properties.
Supplementary information
Supplementary Table 1
Spatial properties of the three experimental CA2 groups.
Rights and permissions
About this article
Cite this article
Donegan, M.L., Stefanini, F., Meira, T. et al. Coding of social novelty in the hippocampal CA2 region and its disruption and rescue in a 22q11.2 microdeletion mouse model. Nat Neurosci 23, 1365–1375 (2020). https://doi.org/10.1038/s41593-020-00720-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00720-5
This article is cited by
-
Ketamine’s rapid and sustained antidepressant effects are driven by distinct mechanisms
Cellular and Molecular Life Sciences (2024)
-
The CLDN5 gene at the blood-brain barrier in health and disease
Fluids and Barriers of the CNS (2023)
-
Dynamic and stable hippocampal representations of social identity and reward expectation support associative social memory in male mice
Nature Communications (2023)
-
Activation of the CA2-ventral CA1 pathway reverses social discrimination dysfunction in Shank3B knockout mice
Nature Communications (2023)
-
Hippocampal representation during collective spatial behaviour in bats
Nature (2023)