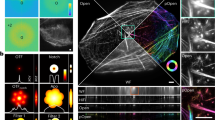
Abstract
With the widespread uptake of two-dimensional (2D) and three-dimensional (3D) single-molecule localization microscopy (SMLM), a large set of different data analysis packages have been developed to generate super-resolution images. In a large community effort, we designed a competition to extensively characterize and rank the performance of 2D and 3D SMLM software packages. We generated realistic simulated datasets for popular imaging modalities—2D, astigmatic 3D, biplane 3D and double-helix 3D—and evaluated 36 participant packages against these data. This provides the first broad assessment of 3D SMLM software and provides a holistic view of how the latest 2D and 3D SMLM packages perform in realistic conditions. This resource allows researchers to identify optimal analytical software for their experiments, allows 3D SMLM software developers to benchmark new software against the current state of the art, and provides insight into the current limits of the field.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Simulated competition datasets are available at http://bigwww.epfl.ch/smlm/challenge2016/, together with the parameters used to generate the data. The ground-truth list of simulated molecule positions for each competition dataset remains secret to allow the software challenge to remain continuously open to new submissions. However, ground-truth data are available for the simulated training datasets. Source data for Figs. 1–4 and for Supplementary Figs. 4–7, 19, 20 and 22 are available online.
Code availability
All software is available at https://github.com/SMLM-Challenge/Challenge2016.
Change history
16 May 2019
In the version of this paper originally published, Figure 4a contained errors that were introduced during typesetting. The bottom 11° ThunderSTORM image is an xz view but was incorrectly labeled as xy, and the low x-axis value in the four line profiles was incorrectly set as –60 instead of –50. These errors have been corrected in the PDF and HTML versions of the paper.
References
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S. T., Girirajan, T. P. K. & Mason, M. D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Holden, S. J., Uphoff, S. & Kapanidis, A. N. DAOSTORM: an algorithm for high- density super-resolution microscopy. Nat. Methods 8, 279–280 (2011).
Huang, F., Schwartz, S. L., Byars, J. M. & Lidke, K. A. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed. Opt. Express 2, 1377–1393 (2011).
Huang, F. et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat. Methods 10, 653–658 (2013).
Sage, D. et al. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods 12, 717–724 (2015).
Huang, B., Jones, S. A., Brandenburg, B. & Zhuang, X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods 5, 1047–1052 (2008).
Shtengel, G. et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl Acad. Sci. USA 106, 3125–3130 (2009).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Babcock, H., Sigal, Y. M. & Zhuang, X. A high-density 3D localization algorithm for stochastic optical reconstruction microscopy. Opt. Nanoscopy 1, 1–10 (2012).
Ovesný, M., Křížek, P., Švindrych, Z. & Hagen, G. M. High density 3D localization microscopy using sparse support recovery. Opt. Express 22, 31263–31276 (2014).
Min, J. et al. 3D high-density localization microscopy using hybrid astigmatic/biplane imaging and sparse image reconstruction. Biomed. Opt. Express 5, 3935–3948 (2014).
Zhang, S., Chen, D. & Niu, H. 3D localization of high particle density images using sparse recovery. Appl. Opt. 54, 7859–7864 (2015).
Juette, M. F. et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529 (2008).
Pavani, S. R. P. et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl Acad. Sci. USA 106, 2995–2999 (2009).
Anonymous. Collaboration through competition. Nat. Methods 11, 695 (2014).
Annibale, P., Vanni, S., Scarselli, M., Rothlisberger, U. & Radenovic, A. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS ONE 6, e22678 (2011).
Li, Y. et al. Real-time 3D single-molecule localization using experimental point spread functions. Nat. Methods 15, 367–369 (2018).
Loot, A., Valdmann, A., Eltermann, M., Kree, M. & Pärs, M. SMolPhot software. BitBucket https://bitbucket.org/ardiloot/ (2016).
Grover, G., DeLuca, K., Quirin, S., DeLuca, J. & Piestun, R. Super-resolution photon-efficient imaging by nanometric double-helix point spread function localization of emitters (SPINDLE). Opt. Express 20, 26681–26695 (2012).
Babcock, H. P. & Zhuang, X. Analyzing single molecule localization microscopy data using cubic splines. Sci. Rep. 7, 552 (2017).
Boyd, N., Schiebinger, G. & Recht, B. The alternating descent conditional gradient method for sparse inverse problems. SIAM J. Optim. 27, 616–639 (2017).
Henriques, R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 (2010).
Takeshima, T., Takahashi, T., Yamashita, J., Okada, Y. & Watanabe, S. A multi-emitter fitting algorithm for potential live cell super-resolution imaging over a wide range of molecular densities. J. Microsc. 271, 266–281 (2018).
Kechkar, A., Nair, D., Heilemann, M., Choquet, D. & Sibarita, J.-B. Real-time analysis and visualization for single-molecule based super-resolution microscopy. PLoS ONE 8, e62918 (2013).
Ovesný, M., Křížek, P., Borkovec, J., Švindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Soubies, E., Blanc-Féraud, L. & Aubert, G. A continuous exact l0 penalty (CEL0) for least squares regularized problem. SIAM J. Imaging Sci. 8, 1607–1639 (2015).
Babcock, H. P., Moffitt, J. R., Cao, Y. & Zhuang, X. Fast compressed sensing analysis for super-resolution imaging using L1-homotopy. Opt. Express 21, 28583–28596 (2013).
Min, J. et al. FALCON: fast and unbiased reconstruction of high-density super-resolution microscopy data. Sci. Rep. 4, 4577 (2014).
Huang, J., Sun, M., Ma, J. & Chi, Y. Super-resolution image reconstruction for high-density three-dimensional single-molecule microscopy. IEEE Trans. Comput. Imaging 3, 763–773 (2017).
Pan, H., Simeoni, M., Hurley, P., Blu, T. & Vetterli, M. LEAP: looking beyond pixels with continuous-space estimation of point sources. Astron. Astrophys. 608, A136 (2017).
Durisic, N., Laparra-Cuervo, L., Sandoval-Álvarez, A., Borbely, J. S. & Lakadamyali, M. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nat. Methods 11, 156–162 (2014).
Chao, J., Ward, E. S. & Ober, R. J. A software framework for the analysis of complex microscopy image data. IEEE Trans. Inf. Technol. Biomed. 14, 1075–1087 (2010).
Martens, K. J. A., Bader, A. N., Baas, S., Rieger, B. & Hohlbein, J. Phasor based single-molecule localization microscopy in 3D (pSMLM-3D): an algorithm for MHz localization rates using standard CPUs. J. Chem. Phys. 148, 123311 (2018).
Marsh, R. J. et al. Artifact-free high-density localization microscopy analysis. Nat. Methods 15, 689–692 (2018).
Ouyang, W., Aristov, A., Lelek, M., Hao, X. & Zimmer, C. Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 36, 460–468 (2018).
Zhang, P. et al. Analyzing complex single-molecule emission patterns with deep learning. Nat. Methods 15, 913–916 (2018).
Boyd, N., Jonas, E., Babcock, H. P. & Recht, B. DeepLoco: fast 3D localization microscopy using neural networks. bioRxiv Preprint at https://www.biorxiv.org/content/10.1101/267096v1 (2018).
Nehme, E., Weiss, L. E., Michaeli, T. & Shechtman, Y. Deep-STORM: super-resolution single-molecule microscopy by deep learning. Optica 5, 458–464 (2018).
Cox, S. et al. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat. Methods 9, 195–200 (2011).
Dertinger, T., Colyer, R., Iyer, G., Weiss, S. & Enderlein, J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI). Proc. Natl Acad. Sci. USA 106, 22287–22292 (2009).
Gustafsson, N. et al. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 7, 12471 (2016).
Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Carlini, L. & Manley, S. Live intracellular super-resolution imaging using site-specific stains. ACS Chem. Biol. 8, 2643–2648 (2013).
Shim, S.-H. et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl Acad. Sci. USA 109, 13978–13983 (2012).
Hanser, B. M., Gustafsson, M. G. L., Agard, D. A. & Sedat, J. W. Phase-retrieved pupil functions in wide-field fluorescence microscopy. J. Microsc. 216, 32–48 (2004).
Izeddin, I. et al. PSF shaping using adaptive optics for three-dimensional single-molecule super-resolution imaging and tracking. Opt. Express 20, 4957–4967 (2012).
McGorty, R., Schnitzbauer, J., Zhang, W. & Huang, B. Correction of depth-dependent aberrations in 3D single-molecule localization and super-resolution microscopy. Opt. Lett. 39, 275–278 (2014).
Hirsch, M., Wareham, R. J., Martin-Fernandez, M. L., Hobson, M. P. & Rolfe, D. J. A stochastic model for electron multiplication charge-coupled devices–from theory to practice. PLoS ONE 8, e53671 (2013).
Basden, A. G., Haniff, C. A. & Mackay, C. D. Photon counting strategies with low-light-level CCDs. Mon. Not. R. Astron. Soc. 345, 985–991 (2003).
Carlini, L., Holden, S. J., Douglass, K. M. & Manley, S. Correction of a depth-dependent lateral distortion in 3D super-resolution imaging. PLoS ONE 10, e0142949 (2015).
Baddeley, D. & Bewersdorf, J. Biological insight from super-resolution microscopy: what we can learn from localization-based images. Annu. Rev. Biochem. 87, 965–989 (2018).
Acknowledgements
The authors acknowledge the following funding sources: a Newcastle University Research Fellowship and a Wellcome Trust and Royal Society Sir Henry Dale Fellowship grant number 206670/Z/17/Z to S.H.; a European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program, Grant Agreement number 692726 to D.S., T.A.P., and M.U.; UK BBSRC grants BB/M022374/1, BB/P027431/1, BB/R000697/1 grant and MRC grants MC-UU-12018/2, MR/K015826/1 to R.H.; ERC grant CoG-724489, CellStructure to J.R.; FranceBioImaging infrastructure ANR-10-INBS-04 to J.-B.S.; National Institutes of Health grant 1R15GM128166-01 to G.M.H.; and NSF SBIR grants 1353638, 1534745 to Double Helix LLC. We thank R. Piestun at University of Colorado for providing double-helix PSF phase mask designs to Double Helix LLC. We thank all the localization microscopy challenge participants for their contribution: H. Babcock (3D-DAOSTORM, Cspline, L1H); F. Hauser (3D STORM Tools); S. Watanabe (3D-WTM,WTM); N. Boyd (ADCG); J. Min, K. Jin and J.C. Ye (ALOHA, FALCON); H. Rouault (B-recs); E. Soubies (CEL0-STORM); A. Speiser, S. Turagas and J. Macke (DECODE); A. von Diezmann, C. Bayas and W.E. Moerner (Easy-DHPSF); T. Vomhof and J. Reichel (FIRESTORM); H. Pan (LEAP); A. Wheeler (Localizer); Z.-l. Huang and Y. Wang (MaLiang); J. Chao, R. Velmurugan, A.V. Abraham and R.J. Ober (MIATool); H. Deschout (mlePALM); T. Pengo (Octane, PeakSelector); Y.-n. Wang (PALMER); A. Herbert (PeakFit); K. Martens and J. Hohlbein (pSMLM-3D); L. Li (QC-STORM); R. Henriques (QuickPALM); G. Tamas and J. Sinko (RainSTORM); S. Wolter and M. Sauer (RapidSTORM); M. Kirchgessner and F. Gruell (SFP Estimator); Y. Li and J. Ries (SMAP); H. Ikoma (SMfit); A. Loot, A. Valdmann, M. Eltermann, M. Kree and M. Pärs (SMolPhot); Y.J. Jung, A. Barsic, R. Piestun and N. Fakhri (SOLAR_STORM); A. Archetti (STORMChaser); M. Ovesny, G. Hagen and P. Krizek (ThunderSTORM); J. Huang (TVSTORM); A. Kechkar, C. Butler and J.-B. Sibarita (WaveTracer) and B. Lelandais (ZOLA-3D). We thank the SMLMS 2016 organizers (S. Manley and A. Radenovic, EPFL) for hosting a localization microscopy challenge special session. We also thank Double Helix and Molecular Devices for sponsoring the SMLMS 2016 special session. The sponsors had no input or influence on the research.
Author information
Authors and Affiliations
Contributions
D.S. and S.H. conceived and coordinated the study. D.S., S.H., T.-A.P., A. Archetti, H.B., S.C., A.W., G.M.H., R.H., T.L., T.P., and J.-B.S. designed the study. S.H., A. Agrawal, R.H., and J.-B.S. collected experimental PSFs. D.S., T.-A.P., S.H., and T.L. wrote simulation code. B.R. shared unpublished software. D.S. generated simulated datasets. J.R. shared experimental STORM data. A.H., J.R., J.C., and R.V. provided feedback and quality control on simulations and analysis methods. T.-A.P. carried out the assessment of software performance. T.-A.P., D.S., and S.H. analyzed and interpreted the results. D.S., H.B., R.O., B.R., G.M.H., J.-B.S., J.R., R.H., M.U., and S.H. directed research. S.H., D.S., and T.-A.P. wrote the manuscript with feedback from all other authors.
Corresponding authors
Ethics declarations
Competing interests
A. Agrawal is an employee of Double Helix LLC, USA.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–23, Supplementary Tables 1–5 and Supplementary Notes 1–5
Rights and permissions
About this article
Cite this article
Sage, D., Pham, TA., Babcock, H. et al. Super-resolution fight club: assessment of 2D and 3D single-molecule localization microscopy software. Nat Methods 16, 387–395 (2019). https://doi.org/10.1038/s41592-019-0364-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0364-4
This article is cited by
-
High-density volumetric super-resolution microscopy
Nature Communications (2024)
-
Mapping super-resolution image quality
Light: Science & Applications (2024)
-
RegiSTORM: channel registration for multi-color stochastic optical reconstruction microscopy
BMC Bioinformatics (2023)
-
Maximum-likelihood model fitting for quantitative analysis of SMLM data
Nature Methods (2023)
-
Quantitatively mapping local quality of super-resolution microscopy by rolling Fourier ring correlation
Light: Science & Applications (2023)