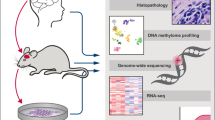
Abstract
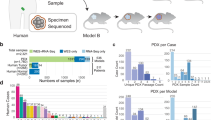
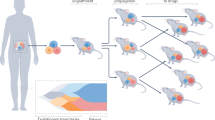
Outcomes of anticancer therapy vary dramatically among patients due to diverse genetic and molecular backgrounds, highlighting extensive intertumoral heterogeneity. The fundamental tenet of precision oncology defines molecular characterization of tumors to guide optimal patient-tailored therapy. Towards this goal, we have established a compilation of pharmacological landscapes of 462 patient-derived tumor cells (PDCs) across 14 cancer types, together with genomic and transcriptomic profiling in 385 of these tumors. Compared with the traditional long-term cultured cancer cell line models, PDCs recapitulate the molecular properties and biology of the diseases more precisely. Here, we provide insights into dynamic pharmacogenomic associations, including molecular determinants that elicit therapeutic resistance to EGFR inhibitors, and the potential repurposing of ibrutinib (currently used in hematological malignancies) for EGFR-specific therapy in gliomas. Lastly, we present a potential implementation of PDC-derived drug sensitivities for the prediction of clinical response to targeted therapeutics using retrospective clinical studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All sequenced data have been deposited in the European Genome-phenome Archive (EGA) under accession EGAS00001002515. Processed data and basic association analysis are publicly available through an interactive web portal (the Cancer-Drug eXplorer (cDx); see URLs).
References
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Weinstein, J. N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Hamburg, M. A. & Collins, F. S. The path to personalized medicine. N. Engl. J. Med. 363, 301–304 (2010).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011).
O’Brien, S. G. et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 348, 994–1004 (2003).
Loeb, L. A. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer 11, 450–457 (2011).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Ceccarelli, M. et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016).
Rubio-Perez, C. et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–396 (2015).
Altman, R. B. Predicting cancer drug response: advancing the DREAM. Cancer Discov. 5, 237–238 (2015).
Geeleher, P., Cox, N. J. & Huang, R. S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 15, R47 (2014).
Lee, J. K. et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 49, 594–599 (2017).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Yates, L. R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 21, 751–759 (2015).
Garnett, M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Shoemaker, R. H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823 (2006).
Basu, A. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013).
Holbeck, S. L., Collins, J. M. & Doroshow, J. H. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 9, 1451–1460 (2010).
Garnett, M. J. & McDermott, U. The evolving role of cancer cell line-based screens to define the impact of cancer genomes on drug response. Curr. Opin. Genet. Dev. 24, 114–119 (2014).
Van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Iorio, F. et al. A landscape of pharmacogenomic interactions in cancer. Cell 166, 740–754 (2016).
Gao, H. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 21, 1318–1325 (2015).
Galli, R. et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021 (2004).
Joo, K. M. et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 3, 260–273 (2013).
Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006).
Lee, J. Y. et al. Patient-derived cell models as preclinical tools for genome-directed targeted therapy. Oncotarget 6, 25619–25630 (2015).
Xie, Y. et al. The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine 2, 1351–1363 (2015).
Kanabur, P. et al. Patient-derived glioblastoma stem cells respond differentially to targeted therapies. Oncotarget 7, 86406–86419 (2016).
Park, Y. H. et al. Role of HER2 mutations in refractory metastatic breast cancers: targeted sequencing results in patients with refractory breast cancer. Oncotarget 6, 32027–32038 (2015).
Lim, S. H. et al. The implication of FLT3 amplification for FLT targeted therapeutics in solid tumors. Oncotarget 8, 3237–3245 (2017).
Yoo, K. H. et al. Genomic alterations in biliary tract cancer using targeted sequencing. Transl. Oncol. 9, 173–178 (2016).
Song, H. N. et al. Molecular characterization of colorectal cancer patients and concomitant patient-derived tumor cell establishment. Oncotarget 7, 19610–19619 (2016).
Suzuki, H. et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015).
Trifonov, V., Pasqualucci, L., Tiacci, E., Falini, B. & Rabadan, R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst. Biol. 7, S2 (2013).
Magi, A. et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 14, R120 (2013).
Abate, F. et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst. Biol. 8, 97 (2014).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Gschwind, A., Fischer, O. M. & Ullrich, A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer 4, 361–370 (2004).
Nakada, M. et al. Aberrant signaling pathways in glioma. Cancers (Basel) 3, 3242–3278 (2011).
Joo, K. M. et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 72, 3828–3838 (2012).
Wen, P. Y., Lee, E. Q., Reardon, D. A., Ligon, K. L. & Alfred Yung, W. K. Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro. Oncol. 14, 819–829 (2012).
Filbin, M. G. et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat. Med. 19, 1518–1523 (2013).
Wen, P. Y. & Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 359, 492–507 (2008).
Ohka, F., Natsume, A. & Wakabayashi, T. Current trends in targeted therapies for glioblastoma multiforme. Neurol. Res. Int. 2012, 878425 (2012).
Puputti, M. et al. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in gliomas. Mol. Cancer Res. 4, 927–934 (2006).
Taylor, T. E., Furnari, F. B. & Cavenee, W. K. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr. Cancer Drug Targets 12, 197–209 (2012).
Snuderl, M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 20, 810–817 (2011).
Szerlip, N. J. et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl Acad. Sci. USA 109, 3041–3046 (2012).
Cloughesy, T. F., Cavenee, W. K. & Mischel, P. S. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 9, 1–25 (2014).
Mellinghoff, I. K. et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 353, 2012–2024 (2005).
Fallahi-Sichani, M., Honarnejad, S., Heiser, L. M., Gray, J. W. & Sorger, P. K. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat. Chem. Biol. 9, 708–714 (2013).
Jang, I. S., Neto, E. C., Guinney, J., Friend, S. H. & Margolin, A. A. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Pac. Symp. Biocomput. 2014, 63–74 (2014).
Huang, S. & Pang, L. Comparing statistical methods for quantifying drug sensitivity based on in vitro dose-response assays. Assay Drug Dev. Technol. 10, 88–96 (2012).
Raub, T. J. et al. Brain exposure of two selective dual CDK4 and CDK6 inhibitors and the antitumor activity of CDK4 and CDK6 inhibition in combination with temozolomide in an intracranial glioblastoma xenograft. Drug Metab. Dispos. 43, 1360–1371 (2015).
Cen, L. et al. p16–Cdk4–Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro. Oncol. 14, 870–881 (2012).
Schroder, L. B. & McDonald, K. L. CDK4/6 inhibitor PD0332991 in glioblastoma treatment: does it have a future? Front. Oncol. 5, 259 (2015).
Nicolau, M., Levine, A. J. & Carlsson, G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc. Natl Acad. Sci. USA 108, 7265–7270 (2011).
Camara, P. G., Rosenbloom, D. I., Emmett, K. J., Levine, A. J. & Rabadan, R. Topological data analysis generates high-resolution, genome-wide maps of human recombination. Cell Syst. 3, 83–94 (2016).
Rizvi, A. H. et al. Applied topology delineates developmental progression with single-cell resolution. Nat. Biotech. (in the press).
Bhattacharya, B. et al. Pharmacologic synergy between dual phosphoinositide-3-kinase and mammalian target of rapamycin inhibition and 5-fluorouracil in PIK3CA mutant gastric cancer cells. Cancer Biol. Ther. 13, 34–42 (2012).
Tapia, O. et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 465, 25–33 (2014).
Ying, J. et al. The expression of the PI3K/AKT/mTOR pathway in gastric cancer and its role in gastric cancer prognosis. OncoTargets Ther. 8, 2427–2433 (2015).
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Yiin, J. J. et al. ZD6474, a multitargeted inhibitor for receptor tyrosine kinases, suppresses growth of gliomas expressing an epidermal growth factor receptor mutant, EGFRvIII, in the brain. Mol. Cancer Ther. 9, 929–941 (2010).
Gao, W. et al. Selective antitumor activity of ibrutinib in EGFR-mutant non-small cell lung cancer cells. J. Natl Cancer. Inst. 106, dju204 (2014).
Byrd, J. C. et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 369, 32–42 (2013).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Garassino, M. C. et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann. Oncol. 22, 235–237 (2011).
Lievre, A. et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 66, 3992–3995 (2006).
Belmont, P. J. et al. Resistance to dual blockade of the kinases PI3K and mTOR in KRAS-mutant colorectal cancer models results in combined sensitivity to inhibition of the receptor tyrosine kinase EGFR. Sci. Signal. 7, ra107 (2014).
Hutchinson, L. Targeted therapies: dasatinib sensitizes KRAS-mutant colorectal cancer tumors to cetuximab. Nat. Rev. Clin. Oncol. 8, 193 (2011).
Ku, B. M. et al. BYL719, a selective inhibitor of phosphoinositide 3-kinase alpha, enhances the effect of selumetinib (AZD6244, ARRY-142886) in KRAS-mutant non-small cell lung cancer. Invest. New Drugs 33, 12–21 (2015).
Jing, J. et al. Comprehensive predictive biomarker analysis for MEK inhibitor GSK1120212. Mol. Cancer Ther. 11, 720–729 (2012).
Infante, J. R. et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 13, 773–781 (2012).
Hatzivassiliou, G. et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature 501, 232–236 (2013).
Blumenschein, G. R. Jr. et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann. Oncol. 26, 894–901 (2015).
Manchado, E. et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–651 (2016).
Yeh, J. J. et al. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol. Cancer Ther. 8, 834–843 (2009).
Sun, C. et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7, 86–93 (2014).
Heinemann, V., Stintzing, S., Kirchner, T., Boeck, S. & Jung, A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat. Rev. 35, 262–271 (2009).
Cui, J., Jiang, W., Wang, S., Wang, L. & Xie, K. Role of Wnt/beta-catenin signaling in drug resistance of pancreatic cancer. Curr. Pharm. Des. 18, 2464–2471 (2012).
Yeung, J. et al. Beta-catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell 18, 606–618 (2010).
Nagaraj, A. B. et al. Critical role of Wnt/beta-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget 6, 23720–23734 (2015).
Ivanescu, A. M., Oprea, M., Turbatu, A., Colita, A. & Lupu, A. R. Ibrutinib, a novel agent in relapsed or refractory chronic lymphocytic leukemia. Maedica (Buchar) 9, 217–218 (2014).
Rushworth, S. A., MacEwan, D. J. & Bowles, K. M. Ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 369, 1277–1278 (2013).
Wang, M. L. et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 369, 507–516 (2013).
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Wang, J., Kribelbauer, J. & Rabadan, R. Network propagation reveals novel genetic features predicting drug response of cancer cell lines. Curr. Bioinform. 11, 8 (2016).
Munarini, N. et al. Altered mammary epithelial development, pattern formation and involution in transgenic mice expressing the EphB4 receptor tyrosine kinase. J. Cell Sci. 115, 25–37 (2002).
Kumar, S. R. et al. Receptor tyrosine kinase EphB4 is a survival factor in breast cancer. Am. J. Pathol. 169, 279–293 (2006).
Yang, N. Y., Pasquale, E. B., Owen, L. B. & Ethell, I. M. The EphB4 receptor-tyrosine kinase promotes the migration of melanoma cells through Rho-mediated actin cytoskeleton reorganization. J. Biol. Chem. 281, 32574–32586 (2006).
Ferguson, B. D. et al. The EphB4 receptor tyrosine kinase promotes lung cancer growth: a potential novel therapeutic target. PLoS ONE 8, e67668 (2013).
Pasquale, E. B. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180 (2010).
Cai, Y., Yan, X., Zhang, G., Zhao, W. & Jiao, S. The predictive value of ERCC1 and p53 for the effect of panobinostat and cisplatin combination treatment in NSCLC. Oncotarget 6, 18997–19005 (2015).
Lee, E. Q. et al. Phase II study of panobinostat in combination with bevacizumab for recurrent glioblastoma and anaplastic glioma. Neuro. Oncol. 17, 862–867 (2015).
Grasso, C. S. et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med. 21, 555–559 (2015).
Taylor, P. et al. REST is a novel prognostic factor and therapeutic target for medulloblastoma. Mol. Cancer Ther. 11, 1713–1723 (2012).
Wang, Z., Qin, G. & Zhao, T. C. HDAC4: mechanism of regulation and biological functions. Epigenomics 6, 139–150 (2014).
Plass, C. et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013).
Kawata, H. et al. Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem. J. 373, 747–757 (2003).
Walkinshaw, D. R. et al. The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J. Biol. Chem. 288, 9345–9362 (2013).
Geng, L. et al. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res. 66, 11298–11304 (2006).
Geng, H. et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J. Biol. Chem. 286, 38095–38102 (2011).
Choi, M. C. et al. A direct HDAC4–MAP kinase crosstalk activates muscle atrophy program. Mol. Cell 47, 122–132 (2012).
Ellis, L. M. & Hicklin, D. J. Resistance to targeted therapies: refining anticancer therapy in the era of molecular oncology. Clin. Cancer Res. 15, 7471–7478 (2009).
Hopper-Borge, E. A. et al. Mechanisms of tumor resistance to EGFR-targeted therapies. Expert Opin. Ther. Targets 13, 339–362 (2009).
Spaans, J. N. & Goss, G. D. Drug resistance to molecular targeted therapy and its consequences for treatment decisions in non-small-cell lung cancer. Front. Oncol. 4, 190 (2014).
Fan, Q. W. et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 24, 438–449 (2013).
Nathanson, D. A. et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343, 72–76 (2014).
Thiessen, B. et al. A phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother. Pharmacol. 65, 353–361 (2010).
Reardon, D. A. et al. Phase I/randomized phase II study of afatinib, an irreversible ErbB family blocker, with or without protracted temozolomide in adults with recurrent glioblastoma. Neuro. Oncol. 17, 430–439 (2015).
Uhm, J. H. et al. Phase II evaluation of gefitinib in patients with newly diagnosed grade 4 astrocytoma: Mayo/North Central Cancer Treatment Group Study N0074. Int. J. Radiat. Oncol. Biol. Phys. 80, 347–353 (2011).
Ritch, P. S., Carroll, S. L. & Sontheimer, H. Neuregulin-1 enhances survival of human astrocytic glioma cells. Glia 51, 217–228 (2005).
Sheng, Q. et al. An activated ErbB3/NRG1 autocrine loop supports in vivo proliferation in ovarian cancer cells. Cancer Cell 17, 298–310 (2010).
Wilson, T. R., Lee, D. Y., Berry, L., Shames, D. S. & Settleman, J. Neuregulin-1-mediated autocrine signaling underlies sensitivity to HER2 kinase inhibitors in a subset of human cancers. Cancer Cell 20, 158–172 (2011).
Dong, X., Fernandez-Salas, E., Li, E. & Wang, S. Elucidation of resistance mechanisms to second-generation ALK inhibitors alectinib and ceritinib in non-small cell lung cancer cells. Neoplasia 18, 162–171 (2016).
Dhomen, N. S., Mariadason, J., Tebbutt, N. & Scott, A. M. Therapeutic targeting of the epidermal growth factor receptor in human cancer. Crit. Rev. Oncog. 17, 31–50 (2012).
Dempke, W. C. & Heinemann, V. Ras mutational status is a biomarker for resistance to EGFR inhibitors in colorectal carcinoma. Anticancer Res. 30, 4673–4677 (2010).
Fan, Q. W. et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res. 67, 7960–7965 (2007).
Yi, Y. W. et al. Inhibition of the PI3K/AKT pathway potentiates cytotoxicity of EGFR kinase inhibitors in triple-negative breast cancer cells. J. Cell. Mol. Med. 17, 648–656 (2013).
Tricker, E. M. et al. Combined EGFR/MEK inhibition prevents the emergence of resistance in EGFR-mutant lung cancer. Cancer Discov. 5, 960–971 (2015).
Huang, M. H. et al. MEK inhibitors reverse resistance in epidermal growth factor receptor mutation lung cancer cells with acquired resistance to gefitinib. Mol. Oncol. 7, 112–120 (2013).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Soreide, K. Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J. Clin. Pathol. 62, 1–5 (2009).
Hajian-Tilaki, K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J. Intern. Med. 4, 627–635 (2013).
Rich, J. N. et al. Phase II trial of gefitinib in recurrent glioblastoma. J. Clin. Oncol. 22, 133–142 (2004).
Lasocki, A., Gaillard, F., Tacey, M., Drummond, K. & Stuckey, S. Multifocal and multicentric glioblastoma: improved characterisation with FLAIR imaging and prognostic implications. J. Clin. Neurosci. 31, 92–98 (2016).
Liu, Q. et al. Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta Neuropathol. 130, 587–597 (2015).
Alshami, J. et al. Afatinib, an irreversible ErbB family blocker, with protracted temozolomide in recurrent glioblastoma: a case report. Oncotarget 6, 34030–34037 (2015).
Ma, D. J. et al. A phase II trial of everolimus, temozolomide, and radiotherapy in patients with newly diagnosed glioblastoma: NCCTG N057K. Neuro. Oncol. 17, 1261–1269 (2015).
Kaley, T. J. et al. Phase II trial of sunitinib for recurrent and progressive atypical and anaplastic meningioma. Neuro. Oncol. 17, 116–121 (2015).
Bang, Y. J. et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010).
Hecht, J. R. et al. Lapatinib in combination with capecitabine plus oxaliplatin in human epidermal growth factor receptor 2-positive advanced or metastatic gastric, esophageal, or gastroesophageal adenocarcinoma: TRIO-013/LOGiC—a randomized phase III trial. J. Clin. Oncol. 34, 443–451 (2016).
Kim, S. T. et al. Impact of genomic alterations on lapatinib treatment outcome and cell-free genomic landscape during HER2 therapy in HER2-positive gastric cancer patients. Ann. Oncol. 29, 1037–1048 (2018).
The Cancer Genome Atlas Research Network.. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Fernandez-Cuesta, L. & Thomas, R. K. Molecular pathways: targeting NRG1 fusions in lung cancer. Clin. Cancer Res. 21, 1989–1994 (2015).
Wu, H. et al. Ibrutinib selectively and irreversibly targets EGFR (L858R, Del19) mutant but is moderately resistant to EGFR (T790M) mutant NSCLC cells. Oncotarget 6, 31313–31322 (2015).
Bernard, S. et al. Activity of ibrutinib in mantle cell lymphoma patients with central nervous system relapse. Blood 126, 1695–1698 (2015).
Jain, P. et al. Early responses predict better outcomes in patients with newly diagnosed chronic myeloid leukemia: results with four tyrosine kinase inhibitor modalities. Blood 121, 4867–4874 (2013).
Louvet, C. et al. Correlation between progression free survival and response rate in patients with metastatic colorectal carcinoma. Cancer 91, 2033–2038 (2001).
Tsimberidou, A. M. & Kurzrock, R. Precision medicine: lessons learned from the SHIVA trial. Lancet Oncol. 16, e579–e580 (2015).
Baras, A., Yu, Y., Filtz, M., Kim, B. & Moskaluk, C. A. Combined genomic and gene expression microarray profiling identifies ECOP as an upregulated gene in squamous cell carcinomas independent of DNA amplification. Oncogene 28, 2919–2924 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 29, (15–21 (2013).
Iyer, M. K., Chinnaiyan, A. M. & Maher, C. A. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 27, 2903–2904 (2011).
Torres-Garcia, W. et al. PRADA: pipeline for RNA sequencing data analysis. Bioinformatics 30, 2224–2226 (2014).
Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999).
Franceschini, A. et al. STRINGv9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013).
Griffith, M. et al. DGIdb: mining the druggable genome. Nat. Methods 10, 1209–1210 (2013).
Kondor, R. I. & Lafferty, J. Diffusion kernels on graphs and other discrete structures. In Proc. 19th International Conference on Machine Learning 8 (Morgan Kaufmann, 2002).
Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67, 20 (2005).
Honigberg, L. A. et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc. Natl Acad. Sci. USA 107, 13075–13080 (2010).
Acknowledgements
This research was supported by a grant of the Korea Health Technology Research and Development project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (HI14C3418). This work has been funded by NIH grants (R01 CA185486, R01 CA179044, U54 CA193313 and U54 209997) and NSF/SU2C/V Foundation Ideas Lab Multidisciplinary Team (PHY-1545805) and Hong Kong RGC grants (N_HKUST601/17 and C6002-17G). The biospecimens for this study were provided by the Samsung Medical Center BioBank.
Author information
Authors and Affiliations
Contributions
J.-K.L., Z.L., J.K.S., S.S. and J.W. are co-first authors. J.-K.L., Z.L., J.K.S., S.S. and J.W. performed the majority of the experiments and analyses. Z.L. and M.B. analyzed the therapeutic landscape of PDCs and pharmacogenomic interactions. D.I.S.R., O.E. and T.C. designed and constructed the cDx interactive webportal. S.W.C., D.-S.K., D.-H.N., S.T.K. and J.L. interpreted the clinical data. J.-K.L., S.S., J.-W.O., M.S., H.J.K., S.H.K., G.H.R. and Y.-J.K. organized and analyzed the drug-screening experiments. Y.J.Shin, H.J.K., Y.J.Seo, M.L., S.Y.K., M.-H.S., J.K., T.L., S.-Y.S., K.-M.K., M.K., J.O.P. and Y.Y. organized and processed the specimens for patient-derived cultures and genome analysis. D.K. and M.L. conducted the animal experiments. J.K.S., H.J.C., I.-H.L., H.S., N.K.D.K., J.S.B. and W.-Y.P. analyzed the genomic profiling. D.-S.K., J.W.C., H.J.S., J.-I.L., J.-W.L., H.-C.K., J.E.L., M.G.C., S.W.S., Y.M.S., J.I.Z. and B.C.J. provided surgical specimens. J.-K.L., Z.L., J.K.S., S.S. and J.W. wrote the manuscript with the feedback from J.L., R.G.W.V., A.I., J.L., R.R. and D.-H.N. J.L., R.R. and D.-H.N. designed and supervised the entire project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15
Supplementary Table 1
Clinical information of the pan-cancer patients included in this study
Supplementary Table 2
CancerSCAN (targeted exome sequencing) gene list
Supplementary Table 3
GliomaSCAN (targeted exome sequencing) gene list
Supplementary Table 4
List of detected genomic alterations (mutation, fusion, copy number variation)
Supplementary Table 5
List of the 60-drug panel
Supplementary Table 6
Sixty-drug library quality control
Supplementary Table 7
Area under the curve (AUC) for the dose–response curve (DRC)
Supplementary Table 8
Half-maximal inhibitory concentration of drug sensitivity
Supplementary Table 9
Cancer-type-specific drug associations
Supplementary Table 10
Topolgoical data analysis of cancer-type-specific drug associations
Supplementary Table 11
Single genomic alteration–drug associations
Supplementary Table 12
Genetic features associated with panobinostat response using dNetFS
Supplementary Table 13
Genetic features associated with EGFR inhibitor response using dNetFS
Supplementary Table 14
Clinical responses in retrospective cases
Rights and permissions
About this article
Cite this article
Lee, JK., Liu, Z., Sa, J.K. et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat Genet 50, 1399–1411 (2018). https://doi.org/10.1038/s41588-018-0209-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0209-6
This article is cited by
-
Inhibition of histone deacetylase 6 destabilizes ERK phosphorylation and suppresses cancer proliferation via modulation of the tubulin acetylation-GRP78 interaction
Journal of Biomedical Science (2023)
-
Patient-derived cell-based pharmacogenomic assessment to unveil underlying resistance mechanisms and novel therapeutics for advanced lung cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
A fair experimental comparison of neural network architectures for latent representations of multi-omics for drug response prediction
BMC Bioinformatics (2023)
-
PRMT3-mediated arginine methylation of IGF2BP1 promotes oxaliplatin resistance in liver cancer
Nature Communications (2023)
-
DeepInsight-3D architecture for anti-cancer drug response prediction with deep-learning on multi-omics
Scientific Reports (2023)