Abstract
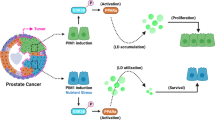
Lipids, either endogenously synthesized or exogenous, have been linked to human cancer. Here we found that PML is frequently co-deleted with PTEN in metastatic human prostate cancer (CaP). We demonstrated that conditional inactivation of Pml in the mouse prostate morphs indolent Pten-null tumors into lethal metastatic disease. We identified MAPK reactivation, subsequent hyperactivation of an aberrant SREBP prometastatic lipogenic program, and a distinctive lipidomic profile as key characteristic features of metastatic Pml and Pten double-null CaP. Furthermore, targeting SREBP in vivo by fatostatin blocked both tumor growth and distant metastasis. Importantly, a high-fat diet (HFD) induced lipid accumulation in prostate tumors and was sufficient to drive metastasis in a nonmetastatic Pten-null mouse model of CaP, and an SREBP signature was highly enriched in metastatic human CaP. Thus, our findings uncover a prometastatic lipogenic program and lend direct genetic and experimental support to the notion that a Western HFD can promote metastasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Wu, J. N., Fish, K. M., Evans, C. P., Devere White, R. W. & Dall’Era, M. A. No improvement noted in overall or cause-specific survival for men presenting with metastatic prostate cancer over a 20-year period. Cancer 120, 818–823 (2014).
Yang, M. et al. Dietary patterns after prostate cancer diagnosis in relation to disease-specific and total mortality. Cancer Prev. Res. (Phila.) 8, 545–551 (2015).
Grönberg, H. Prostate cancer epidemiology. Lancet 361, 859–864 (2003).
Markozannes, G. et al. Diet, body size, physical activity and risk of prostate cancer: an umbrella review of the evidence. Eur. J. Cancer 69, 61–69 (2016).
Zhu, Y. et al. Dietary patterns and colorectal cancer recurrence and survival: a cohort study. BMJ Open 3, e002270 (2013).
Ittmann, M. et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 73, 2718–2736 (2013).
Steck, P. A. et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 15, 356–362 (1997).
Li, J. et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997).
Yoshimoto, M. et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet. Cytogenet. 169, 128–137 (2006).
Sircar, K. et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J. Pathol. 218, 505–513 (2009).
Han, B. et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod. Pathol. 22, 1083–1093 (2009).
Lunardi, A. et al. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat. Genet. 45, 747–755 (2013).
Beltran, H. et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur. Urol. 63, 920–926 (2013).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Taylor, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Song, M. S., Salmena, L. & Pandolfi, P. P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell. Biol. 13, 283–296 (2012).
Ma, X. et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 65, 5730–5739 (2005).
Trotman, L. C. et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, E59 (2003).
Svensson, R. U. et al. Slow disease progression in a C57BL/6 pten-deficient mouse model of prostate cancer. Am. J. Pathol. 179, 502–512 (2011).
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).
Ding, Z. et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273 (2011).
Gao, H. et al. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc. Natl. Acad. Sci. USA 103, 14477–14482 (2006).
Kinkade, C. W. et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J. Clin. Invest. 118, 3051–3064 (2008).
Wang, X. S. et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 1, 35–43 (2011).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Palanisamy, N. et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat. Med. 16, 793–798 (2010).
Moul, J. W., Friedrichs, P. A., Lance, R. S., Theune, S. M. & Chang, E. H. Infrequent RAS oncogene mutations in human prostate cancer. Prostate. 20, 327–338 (1992).
Gumerlock, P. H., Poonamallee, U. R., Meyers, F. J. & deVere White, R. W. Activated ras alleles in human carcinoma of the prostate are rare. Cancer Res. 51, 1632–1637 (1991).
Barbieri, C. E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012).
Wang, J. et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 72, 4765–4776 (2012).
Mulholland, D. J. et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 72, 1878–1889 (2012).
Walker, E. J. et al. Monoallelic expression determines oncogenic progression and outcome in benign and malignant brain tumors. Cancer Res. 72, 636–644 (2012).
Trotman, L. C. et al. Identification of a tumour suppressor network opposing nuclear Akt function. Nature 441, 523–527 (2006).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Serra, V. et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 30, 2547–2557 (2011).
Carracedo, A. et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Invest. 118, 3065–3074 (2008).
Wang, Z. G. et al. Role of PML in cell growth and the retinoic acid pathway. Science 279, 1547–1551 (1998).
Swinnen, J. V. et al. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int. J. Cancer 98, 19–22 (2002).
Baenke, F., Peck, B., Miess, H. & Schulze, A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6, 1353–1363 (2013).
Goldstein, J. L. & Brown, M. S. A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161, 161–172 (2015).
Porstmann, T. et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236 (2008).
Düvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Trevino, M. B. et al. Perilipin 5 regulates islet lipid metabolism and insulin secretion in a cAMP-dependent manner: implication of its role in the postprandial insulin secretion. Diabetes 64, 1299–1310 (2015).
Breitkopf, S. B., Yuan, M., Helenius, K. P., Lyssiotis, C. A. & Asara, J. M. Triomics analysis of imatinib-treated myeloma cells connects kinase inhibition to RNA processing and decreased lipid biosynthesis. Anal. Chem. 87, 10995–11006 (2015).
Breitkopf, S. B. et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13, 30 (2017).
Shroff, E. H. et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc. Natl. Acad. Sci. USA 112, 6539–6544 (2015).
Eberlin, L. S. et al. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc. Natl. Acad. Sci. USA 111, 10450–10455 (2014).
Perry, R. H. et al. Characterization of MYC-induced tumorigenesis by in situ lipid profiling. Anal. Chem. 85, 4259–4262 (2013).
Nomura, D. K. et al. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140, 49–61 (2010).
Rysman, E. et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 70, 8117–8126 (2010).
Hilvo, M. et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 71, 3236–3245 (2011).
Kotzka, J. et al. Insulin-activated Erk-mitogen-activated protein kinases phosphorylate sterol regulatory element-binding protein-2 at serine residues 432 and 455 in vivo. J. Biol. Chem. 279, 22404–22411 (2004).
Abidi, P., Zhang, F., Li, C. & Liu, J. Blockage of the ERK signaling pathway abrogates the SCAP ligand-induced transcriptional activation of the LDL receptor gene in HepG2 cells. Int. J. Mol. Med. 16, 779–785 (2005).
Ma, L., Chen, Z., Erdjument-Bromage, H., Tempst, P. & Pandolfi, P. P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121, 179–193 (2005).
Roux, P. P., Ballif, B. A., Anjum, R., Gygi, S. P. & Blenis, J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 101, 13489–13494 (2004).
Williams, K. J. et al. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 73, 2850–2862 (2013).
Li, X., Chen, Y. T., Hu, P. & Huang, W. C. Fatostatin displays high antitumor activity in prostate cancer by blocking SREBP-regulated metabolic pathways and androgen receptor signaling. Mol. Cancer Ther. 13, 855–866 (2014).
Kamisuki, S. et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 16, 882–892 (2009).
Lin, J. et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell 120, 261–273 (2005).
Unger, R. H. Lipotoxic diseases. Annu. Rev. Med. 53, 319–336 (2002).
Buettner, R. et al. Defining high-fat-diet rat models: metabolic and molecular effects of different fat types. J. Mol. Endocrinol. 36, 485–501 (2006).
Lane, S., Zhou, S., Pan, T., Dai, Q. & Liu, H. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21, 6418–6428 (2001).
Pascual, G. et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45 (2017).
Nieman, K. M. et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498–1503 (2011).
Park, H. et al. A high-fat diet increases angiogenesis, solid tumor growth, and lung metastasis of CT26 colon cancer cells in obesity-resistant BALB/c mice. Mol. Carcinog. 51, 869–880 (2012).
Kim, E. J. et al. Dietary fat increases solid tumor growth and metastasis of 4T1 murine mammary carcinoma cells and mortality in obesity-resistant BALB/c mice. Breast. Cancer Res. 13, R78 (2011).
Sonn, G. A., Aronson, W. & Litwin, M. S. Impact of diet on prostate cancer: a review. Prostate Cancer Prostatic Dis. 8, 304–310 (2005).
Kwon, O. J., Zhang, B., Zhang, L. & Xin, L. High fat diet promotes prostatic basal-to-luminal differentiation and accelerates initiation of prostate epithelial hyperplasia originated from basal cells. Stem Cell Res. 16, 682–691 (2016).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 (2007).
Ros, S. et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2, 328–343 (2012).
Gurrieri, C. et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J. Natl. Cancer Inst. 96, 269–279 (2004).
Tilch, E. et al. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res. Treat. 143, 385–392 (2014).
Chen, M. et al. Deregulated PP1α phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat. Commun. doi:10.1038/s41467-017-02272-y (2017).
Bernardi, R. et al. PML inhibits HIF-1α translation and neoangiogenesis through repression of mTOR. Nature 442, 779–785 (2006).
Das, S. K., Mondal, A. K. & Elbein, S. C. Distinct gene expression profiles characterize cellular responses to palmitate and oleate. J. Lipid Res. 51, 2121–2131 (2010).
Trotman, L. C. et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–156 (2007).
Mehlem, A., Hagberg, C. E., Muhl, L., Eriksson, U. & Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 8, 1149–1154 (2013).
Lukacs, R. U., Goldstein, A. S., Lawson, D. A., Cheng, D. & Witte, O. N. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc. 5, 702–713 (2010).
Acknowledgements
We thank all the members of the laboratory of P.P.P. for critical comments, and L. Southwood and E. Stack for editing the manuscript. We are grateful to G. Augusto dos Santos for insightful discussions. We thank the BIDMC Histology Core facility and M. Yuan in the BIDMC Mass Spectrometry Core for help with the lipidomics experiments. M.C. was supported in part by a DOD Prostate Cancer Research Program (PCRP) Postdoctoral Training Award. This work was supported by NIH grants R01CA142780, R01CA142874 and R35CA197529 to P.P.P. This work was also partially supported by NIH grants P01CA120964 and R35CA197459 to J.M.A.
Author information
Authors and Affiliations
Contributions
M.C., E.G.-B., Y.-R.L., J.M.K., A.V.M., K.A.W., C.N., J.F., M.D.P., M.S.D. and M.C.-M. performed the experiments. M.C. and P.P.P. conceived and designed the experiments. C.C.-C. and P.P.P. supervised the study. K.S. and X.-S.L. generated Pmlflox/flox mice. J.Z. performed all bioinformatic analyses. J.M.A. and S.B.B. performed lipidomic analyses. J.T.-F. performed TMA analyses of patient samples. S.S. and R.T.B. conducted pathology analyses of mouse tissues. M.C., J.Z., E.G.-B., M.C.-M. and P.P.P. analyzed the data. M.C., L.M., J.G.C. and P.P.P. wrote the manuscript. All authors critically discussed the results and the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Co-loss of PTEN and PML expression in advanced and metastatic human CaP.
(a) Bar graph showing the percentage of co-deletion of PTEN with 58 high-confidence TSGs32 in the Grasso et al. dataset of the mCRPC samples14, respectively (4 out of 62 TSGs from the Walker et al gene list, data not available). The genes co-deleted with PTEN only in metastatic disease among the top 25 TSGs are highlighted in Red. (b,c) Bar graph showing the percentage of deletion of PTEN or PML (b), or PTEN and PML (c) in mCRPC samples from the Robinson et al. dataset25. (d) Representative homozygous or hemizygous focal PML deletion from the Robinson et al. dataset (38% (17/45) of PML deletion was focal)25. Copy number plots with x-axis representing chromosomal 15q and y-axis referring to copy number level. Red open circle indicates genomic position of PML. (e) Representative IHC staining of PTEN or PML showing examples of low, medium and high staining. Scale bar, 50 μm. (f) Table showing the significant correlation of co-loss of PTEN and PML protein expression during the disease progression. The number of cases in each expression category was listed together with Gleason score. (g-j) Overall survival curves for CaP patients after radical prostatectomy based on the expression of PML protein (g), the expression of PTEN protein (h), Gleason score (i), or pathologic stage (j). In f, Pearson’s chi-squared test was used to determine significance.
Supplementary Figure 2 Generation of Pmlflox/flox mice.
(a) Schematic map of the WT Pml locus (top), targeting vector (upper middle) and predicted targeted allele (lower middle) and floxed allele (bottom). The Pml genomic sequence was cloned and inserted into the pEZ-LOX-FRT-DT vector. Black triangles mark the location of loxP sites that were utilized to excise the exon 2. Blue triangles mark the location of FRT sites that were utilized to excise the neomycin resistant cassette. The probes for Southern blot analysis are indicated (5′ and 3′ probes). BamHI digestion of genomic DNA from targeted ES cells was use to distinguish WT and targeted allele. BamHI (B), ScaI (S), NotI (N). (b) Southern blot analysis of recombined ES cell clones after digestion with BamHI and hybridization with the 5’ probe (upper panel) and the neomycin (lower panel) probe. ES cell clones with corrected homologous recombination are highlighted in red. (c) Southern blot analysis of tail DNA from F2 mice after digestion with BamHI and hybridization with the 5′ probe (top), 3′ probe and neomycine probe (bottom). The mice with deletion of neomycin resistant cassette are highlighted in red.
Supplementary Figure 3 Pml loss drives MAPK reactivation and metastatic progression in Pten-null CaP.
(a) IHC staining for Pml in the VP tissues from WT, Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice at 12 weeks of age. (b) H&E and IHC staining of the DLP tissues from Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice at 20 weeks of age. Note that tumors in Ptenpc−/−Pmlpc−/− mice acquired invasive feature. Invasiveness was confirmed by the absence of SM-α-actin staining along with high level of Ki67 staining in the cancer cells. Arrows indicate invasive cancer. (c) Higher magnification of Ptenpc−/−Pmlpc−/− tumors at 13 months of age showing predominate adenocarcinoma (arrows) in the presence of focal features of sarcomatoid carcinoma with high-grade pleomorphic spindle cells (arrowheads). (d) Immunoblot (IB) analysis of tissue lysates for Pml from a WT mouse. (e) H&E-stained low-grade PIN in the VP and DLP tissues from a Pmlpc−/− mouse at 12 months of age. Insets show crowding cells with large nuclei. (f) H&E and IHC staining of lumbar lymph node metastases from three Ptenpc−/−Pmlpc−/− mice. Arrows indicate metastases. (g) IHC staining for phosphor-ERK in the DLP tissues from three pairs of Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice at 12 weeks of age. (h) IB analysis of the DLP tissue lysates from WT, Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice at 12 weeks of age. (i) IHC staining for Pml and phosphor-ERK in the DLP tissues from a Ptenpc−/−Pmlpc+/− mouse at 12 weeks of age. Arrows indicate areas with lower level of Pml, but higher level of p-ERK. Arrowheads indicate areas with higher level of Pml, but lower level of p-ERK. Scale bars in all panels, 50μm. Uncropped images in d and h are shown in Supplementary Fig. 7.
Supplementary Figure 4 Transcriptome and lipidomics profiling of WT, Ptenpc−/− and Ptenpc−/−Pmlpc−/− prostates.
(a) Representative H&E staining of DLP from WT, Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice at 12 weeks of age. Scale bar, 50μm. (b) Heat map of the SREBP targets in the microarray analysis of prostate tissues from the three genotypes of mice. (c-f) GSEA enrichment plot for the targets of LXR, ChREBP, PPARγ, and USF. The up- to down-regulated genes from the ranked gene list were analysed with the GSEA algorithm for enrichment of all gene sets in MSigDB among WT, Ptenpc−/− and Ptenpc−/−Pmlpc−/− prostates. (g,h) The relative intensity of all the identifiable 35 lipid classes (g) or the 30 most abundant fatty acyl chains (h) in prostate tissues from the three genotypes of mice. (i) Heat map of the top 70 most regulated lipid ions in prostate tissues from the three genotypes of mice. (j) The validation of the expression changes of hypoxia-induced target genes by the qPCR among WT, Ptenpc−/− and Ptenpc−/−Pmlpc−/− prostates. Data shown in g, h and j are mean ± s.e.m.
Supplementary Figure 5 SREBP-dependent lipogenesis is critical for PML-loss-induced CaP growth and metastasis.
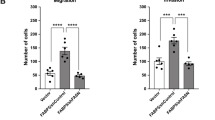
(a,b) Representative images and quantitation of migrated and invaded PC3 cells transfected with siRNA against PML or/and SREBP-1 (a), or LNCaP cells transfected siRNA against SREBP-2 (b), in the migration and invasion assays. CaP cells were transfected with control or indicated siRNA for 48 hrs. PC3 cells were then subjected to 24-hr migration and invasion assay, while LNCaP cells were subjected to 24-hr migration and 48-hr invasion assay. (c) H&E and IHC staining of metastases in the lumbar lymph node of two vehicle-treated Ptenpc−/−Pmlpc−/− mice. Arrows indicate metastases. In a and b, the results of one representative experiment are shown (n = 3). Data are from three independent cultures (4 fields per insert). Data shown are mean ± s.e.m. Student′s t-test (two-tailed) was used to determine significance. Scale bars in all panels, 50 μm.
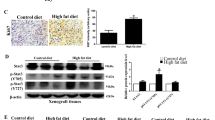
Supplementary Figure 6 A HFD drives metastatic progression in mouse models of CaP and increases lipid abundance in prostate tumors.
(a,b) H&E and IHC staining of metastases in the lung of a representative Ptenpc−/−Pmlpc−/− mouse (a) or a Ptenpc−/− mouse (b). Arrows indicate metastases. (c) The survival analysis of Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice upon 3-month HFD feeding beginning at 12 months of age. (d,e) The relative intensity of all the identifiable 36 lipid classes (d) or the 30 most abundant fatty acyl chains (e) in prostate tissues from chow- or HFD- fed Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice. (f) The ORO staining of vehicle or dietary lipids treated PC3 cells. (g) Representative images and quantitation of migrated or invaded PC3 cells in the migration and invasion assay. PC3 cells were pretreated with BSA, 2% lipid mixture, BSA-conjugate palmitic acid or oleic acid for 7 days, then subjected to 24-hr migration and invasion assay. (h) The serum testosterone levels in chow- or HFD- fed Ptenpc−/− and Ptenpc−/−Pmlpc−/− mice at 14–15 months of age. In g, the results of one representative experiment are shown (n = 5). Data are from three independent cultures (4 fields per insert). Data shown in d, e, g and h are mean ± s.e.m. Student′s t-test (two-tailed) was used to determine significance. Scale bars in all panels, 50 μm.
Supplementary Figure 7
Uncropped scans for the Western blot data
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 8, 9 and 13–16
Supplementary Table 1
Summary of TMA Results and Clinical Information
Supplementary Table 2
The Microarray Results
Supplementary Table 3
GO and GSEA analysis of Microarray Data.
Supplementary Table 4
The Nomenclature of Lipids
Supplementary Tables 5–7
Lipidomics Data from Mouse Prostates at 12 Weeks of Age
Supplementary Tables 10–12
Lipidomics Data from Chow- or HFD-fed Mouse Prostates
Rights and permissions
About this article
Cite this article
Chen, M., Zhang, J., Sampieri, K. et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet 50, 206–218 (2018). https://doi.org/10.1038/s41588-017-0027-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-017-0027-2
This article is cited by
-
Effects of dietary intervention on human diseases: molecular mechanisms and therapeutic potential
Signal Transduction and Targeted Therapy (2024)
-
Lipid metabolic reprogramming in tumor microenvironment: from mechanisms to therapeutics
Journal of Hematology & Oncology (2023)
-
Histone H2A Lys130 acetylation epigenetically regulates androgen production in prostate cancer
Nature Communications (2023)
-
Involvement of the pro-oncogenic enzyme fatty acid synthase in the hallmarks of cancer: a promising target in anti-cancer therapies
Oncogenesis (2023)
-
Physiological and pathological roles of lipogenesis
Nature Metabolism (2023)