Abstract
Red light penetrates deep into mammalian tissues and has low phototoxicity, but few optogenetic tools that use red light have been developed. Here we present MagRed, a red light–activatable photoswitch that consists of a red light–absorbing bacterial phytochrome incorporating a mammalian endogenous chromophore, biliverdin and a photo-state-specific binder that we developed using Affibody library selection. Red light illumination triggers the binding of the two components of MagRed and the assembly of split-proteins fused to them. Using MagRed, we developed a red light–activatable Cre recombinase, which enables light-activatable DNA recombination deep in mammalian tissues. We also created red light–inducible transcriptional regulators based on CRISPR–Cas9 that enable an up to 378-fold activation (average, 135-fold induction) of multiple endogenous target genes. MagRed will facilitate optogenetic applications deep in mammalian organisms in a variety of biological research areas.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the article and its Supplementary Information. The source data for the main figures and extended data figures are provided as Source Data files. A crystal structure of Affibody (Protein Data Bank accession code 2M5A) was used to depict a schematic representation of a binding partner candidate in Fig. 1b. The read counts for all screening data are available on the DDBJ Sequence Read Archive, accession numbers DRR243933 and DRR243934. Source data are provided with this paper.
Code availability
The code for analysis of the read counts for all screening data has been deposited on GitHub (https://github.com/Kazushi40/NGS_analysis).
References
Kennedy, M. J. et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 (2010).
Guntas, G. et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl Acad. Sci. USA 112, 112–117 (2015).
Kawano, F., Suzuki, H., Furuya, A. & Sato, M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 6, 6256 (2015).
Zhang, K. & Cui, B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 33, 92–100 (2015).
Losi, A., Gardner, K. H. & Möglich, A. Blue-light receptors for optogenetics. Chem. Rev. 118, 10659–10709 (2018).
Aoki, K. et al. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell 52, 529–540 (2013).
Nihongaki, Y., Yamamoto, S., Kawano, F., Suzuki, H. & Sato, M. CRISPR–Cas9-based photoactivatable transcription system. Chem. Biol. 22, 169–174 (2015).
Nihongaki, Y. et al. CRISPR–Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 14, 963–966 (2017).
Nihongaki, Y., Kawano, F., Nakajima, T. & Sato, M. Photoactivatable CRISPR–Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760 (2015).
Nihongaki, Y., Otabe, T., Ueda, Y. & Sato, M. A split CRISPR–Cpf1 platform for inducible genome editing and gene activation. Nat. Chem. Biol. 15, 882–888 (2019).
Kawano, F., Okazaki, R., Yazawa, M. & Sato, M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 12, 1059–1064 (2016).
Jung, H. et al. Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat. Commun. 10, 314 (2019).
Lee, D. et al. Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat. Methods 14, 495–503 (2017).
Pu, J., Zinkus-Boltz, J. & Dickinson, B. C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 13, 432–438 (2017).
Yu, D. et al. Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins. Nat. Methods 16, 1095–1100 (2019).
Liu, Q. et al. A photoactivatable botulinum neurotoxin for inducible control of neurotransmission. Neuron 101, 863–875 (2019).
Ziegler, T. & Möglich, A. Photoreceptor engineering. Front. Mol. Biosci. 2, 30 (2015).
Weissleder, R. A clearer vision for in vivo imaging biological imaging. Nat. Biotechnol. 19, 316–317 (2001).
Shimizu-Sato, S., Huq, E., Tepperman, J. M. & Quail, P. H. A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044 (2002).
Levskaya, A., Weiner, O. D., Lim, W. A. & Voigt, C. A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009).
Reichhart, E., Ingles-Prieto, A., Tichy, A. M., McKenzie, C. & Janovjak, H. A phytochrome sensory domain permits receptor activation by red light. Angew. Chem. Int. Ed. 55, 6339–6342 (2016).
Zhou, Y. et al. A small and highly sensitive red/far-red optogenetic switch for applications in mammals. Nat. Biotechnol. 40, 262–272 (2022).
Kohchi, T., Kataoka, H. & Linley, P. J. Biosynthesis of chromophores for phytochrome and related photoreceptors. Plant Biotechnol. 22, 409–413 (2005).
Wu, J. et al. A non-invasive far-red light-induced split-Cre recombinase system for controllable genome engineering in mice. Nat. Commun. 11, 3708 (2020).
Yu, Y. et al. Engineering a far-red light-activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci. Adv. 6, eabb1777 (2020).
Kaberniuk, A., Shemetov, A. A. & Verkhusha, V. V. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat. Methods 13, 591–597 (2016).
Redchuk, T. A., Omelina, E. S., Chernov, K. G. & Verkhusha, V. V. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat. Chem. Biol. 13, 633–639 (2017).
Takala, H. et al. Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 (2014).
Wagner, J. R. et al. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 283, 12212–12226 (2008).
Takala, H., Lehtivuori, H., Hammarén, H., Hytönen, V. P. & Ihalainen, J. A. Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry 53, 7076–7085 (2014).
Nord, K. et al. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat. Biotechnol. 15, 772–777 (1997).
Paulmurugan, R. & Gambhir, S. S. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein–protein interactions. Anal. Chem. 79, 2346–2353 (2007).
Taslimi, A. et al. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 12, 425–430 (2016).
Yao, S. et al. RecV recombinase system for in vivo targeted optogenomic modifications of single cells or cell populations. Nat. Methods 17, 422–429 (2020).
Morikawa, K. et al. Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat. Commun. 11, 2141 (2020).
Sternberg, N. & Hamilton, D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150, 467–486 (1981).
Gierut, J. J., Jacks, T. E. & Haigis, K. M. Strategies to achieve conditional gene mutation in mice. Cold Spring Harb. Protoc. 2014, 339–349 (2014).
Meinke, G., Bohm, A., Hauber, J., Pisabarro, M. T. & Buchholz, F. Cre recombinase and other tyrosine recombinases. Chem. Rev. 116, 12785–12820 (2016).
Kim, H., Kim, M., Im, S.-K. & Fang, S. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 34, 147–159 (2018).
Huang, Z. et al. Creating red light-switchable protein dimerization systems as genetically encoded actuators with high specificity. ACS Synth. Biol. 9, 3322–3333 (2020).
Yen, S. T. et al. CreLite: an optogenetically controlled Cre/loxP system using red light. Dev. Dyn. 249, 1394–1403 (2020).
Hochrein, L., Mitchell, L. A., Schulz, K., Messerschmidt, K. & Mueller-Roeber, B. L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast. Nat. Commun. 9, 1931 (2018).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Dagliyan, O. et al. Computational design of chemogenetic and optogenetic split proteins. Nat. Commun. 9, 4042 (2018).
Dolberg, T. B. et al. Computation-guided optimization of split protein systems. Nat. Chem. Biol. 17, 531–539 (2021).
Legué, E. & Joyner, A. L. Genetic fate mapping using site-specific recombinases. Methods Enzymol. 477, 153–181 (2010).
Pei, W. et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017).
Fenno, L. E. et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11, 763–772 (2014).
Weinberg, B. H. et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol. 35, 453–462 (2017).
Takao, T. et al. Establishment of a tTA-dependent photoactivatable Cre recombinase knock-in mouse model for optogenetic genome engineering. Biochem. Biophys. Res. Commun. 526, 213–217 (2020).
Nakamura, M., Gao, Y., Dominguez, A. A. & Qi, L. S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 23, 11–22 (2021).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Müller, K. et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 41, e77 (2013).
Mukougawa, K., Kanamoto, H., Kobayashi, T., Yokota, A. & Kohchi, T. Metabolic engineering to produce phytochromes with phytochromobilin, phycocyanobilin, or phycoerythrobilin chromophore in Escherichia coli. FEBS Lett. 580, 1333–1338 (2006).
Ohashi, H., Shimizu, Y., Ying, B. W. & Ueda, T. Efficient protein selection based on ribosome display system with purified components. Biochem. Biophys. Res. Commun. 352, 270–276 (2007).
Wang, H. et al. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat. Methods 13, 755–758 (2016).
Skládal, P., Minunni, M., Mascini, M., Kolář, V. & Fránek, M. Characterization of monoclonal antibodies to 2,4-dichlorophenoxyacetic acid using a piezoelectric quartz crystal microbalance in solution. J. Immunol. Methods 176, 117–125 (1994).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Acknowledgements
We thank Y. Aono for support in the analysis of biochemical experiments. This work was supported by a CREST grant (JPMJCR1653) from the Japan Science and Technology Agency (to R.N., M.Y. and M.S.); a project grant from the Kanagawa Institute of Industrial Science and Technology; and the UTEC-UTokyo FSI Research Grant Program to M.S. Y. Kuwasaki was supported by KAKENHI-PROJECT-20J14492 for the Japan Society for the Promotion of Science (JSPS) Doctoral Course Students Research Fellow. K.S. was supported by Grant-in-Aid 18J01772 for the JSPS Research Fellow.
Author information
Authors and Affiliations
Contributions
M.S. conceived the project and provided supervision. G.Y. developed the synthetic binder, with support from Y.S., and performed preliminary tetR-tetO experiments. R.N., K.M., K.F. and Y. Kuwasaki measured absorption spectra of DrBphP and RpBphP1. Y. Kuwasaki, S.Y. and K.M. performed biochemical study using QCM. Y. Kuwasaki and M.N. performed split-fluc reassembly assays. Y. Kakihara and Y. Kuwasaki performed experiments of tetR-tetO. Y. Kuwasaki, T.O. and Y. Kakihara performed experiments of Red-CPTS, with help from T.N. Y. Kuwasaki, M.N., K.S., R.B. and M.Y. performed experiments of RedPA-Cre. Y. Kuwasaki, K.S. and M.S. wrote the manuscript and prepared the figures. R.B. and M.Y. edited the manuscript. All authors checked and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Zhen Gu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
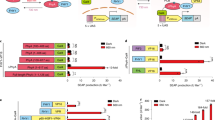
Extended Data Fig. 1 Effect of additional BV supplementation on the three different CPTS designs.
(a-c) Mean bioluminescence intensities (from three independent biological samples) of CPTS designs based on RpBphP1-PpsR2 (a), RpBphP1-QPAS1 (b), and MagRed (c) were plotted. The designs of each configuration (#1-8) were shown in Fig. 3b, c. P values are indicated above the bars. (N.S., not significant P > 0.05; **P < 0.01; BV minus vs. plus using two-tailed Wilcoxon matched-pairs signed rank test).
Extended Data Fig. 2 Red-CPTS can be actively switched off using 800-nm light illumination.
(a-d) After the pre-activation of Red-CPTS by the 660-nm illumination for 12 h, the cells were further incubated under the 660-nm illumination condition (a) or switched to the dark condition (b) or the 800-nm illumination condition (c). As a control, the sample was incubated under the dark condition throughout the experiment (d). ASCL1 mRNA levels were measured at 0 h, 0.75 h, 1.5 h and 3.0 h as shown in the figure. Box plots show the median (center line), first and third quartiles (box edges), 1× the SD (whiskers), and individual data points. (n = 6 biologically independent samples, mean ± s.d.).
Extended Data Fig. 3 Comparison of Red-CPTS and RpBphP1-PspR2/QPAS1-based CPTS at different illumination intensities.
(a-c) Fluc reporter gene activation by Red-CPTS (a), RpBphP1-PpsR2-based CPTS (b), and RpBphP1-QPAS1-based CPTS (c) at various illumination intensities. RpBphP1-PpsR2/QPAS1-based CPTS has configuration #3 shown in Fig. 3b, c. Experimental conditions are same as those in Fig. 3c, d except for the illumination intensities. Ratios of the mean bioluminescence intensity under the red light condition (red bar) to that under the dark condition (gray bar) are depicted above the bars. Bar data are shown as the mean ± s.d. from four biological replicates. Dots represent individual data points. (d) Left: Comparison of the Light/Dark contrasts between Red-CPTS and RpBphP1-PpsR2/QPAS1-based CPTS at different illumination intensities. Right: The cropped data with the mean Light/Dark contrast of 0 to 3.
Extended Data Fig. 4 Dependence of Red-CPTS activity on the duration of ON time of the red light illumination.
(a-c) Fluc reporter gene activation by Red-CPTS (a), RpBphP1-PpsR2 (b), and RpBphP1-QPAS1 (c) with various durations of ON time of the red light illumination. RpBphP1-PpsR2/QPAS1-based CPTS has configuration #3 shown in Fig. 3b, c. Experimental conditions are the same as those in Fig. 3c, d except for the illumination cycle. Ratios of the mean bioluminescence intensity under the red light condition (red bar) to that under the dark condition (gray bar) are depicted above the bars. Bar data are shown as the mean ± s.d. from four biological replicates. Dots represent individual data points. (d) Left: Comparison of the Light/Dark contrasts between Red-CPTS and RpBphP1-PpsR2/QPAS1-based CPTS at various durations of ON time of the red light illumination. Right: The cropped data with the mean Light/Dark contrast of 0 to 3.
Extended Data Fig. 5 Dependence of Red-CPTS activity on the duration of OFF time of the red light illumination.
(a-c) Fluc reporter gene activation by Red-CPTS (a), RpBphP1-PpsR2 (b), and RpBphP1-QPAS1 (c) with various durations of OFF time of the red light illumination. RpBphP1-PpsR2/QPAS1-based CPTS has configuration #3 shown in Fig. 3b, c. Experimental conditions were the same as those in Fig. 3c, d except for the illumination cycle. Ratios of the mean bioluminescence intensity under the red light condition (red bar) to that under the dark condition (gray bar) are depicted above the bars. Bar data are shown as the mean ± s.d. from four biological replicates. Dots represent individual data points. (d) Left: Comparison of the Light/Dark contrasts between Red-CPTS and RpBphP1-PpsR2/QPAS1-based CPTS at various durations of OFF time of the red light illumination. Right: The cropped data with the mean Light/Dark contrast of 0 to 3.
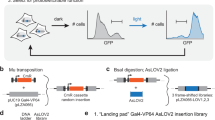
Extended Data Fig. 6 Comparison of RedPA-Cre with the existing red light-responsive recombinase systems.
For the BV/PCB (+) conditions, HEK 293T cells were plated at 2.0 × 104 cells per well in a 96-well black-wall plate in the presence of 25 μM BV (for RedPA-Cre and FISC system) and 20 μM PCB (for CreLite and L-SCRaMbLE), respectively. For the BV/PCB(−) conditions, the procedures for plating were identical to those described above except for the chromophore supplementation. Plasmid amount used for each experiment is described below the graph. Especially, because a previous study has revealed that FISC system shows the highest recombination efficiency when Cre N-fragment (pXY169), Cre C-fragment (pXY177) and red light-responsive activator (pXY137) were transfected at 1:1:10 ratio, we additionally tested this transfection condition for FISC system. Following experimental procedures are the same as those in Fig. 5c, d. Ratios of the mean bioluminescence intensity under the red light condition (red bar) to that under the dark condition (gray bar) are depicted above the bars. Bar data are shown as the mean ± s.d. from four biological replicates. Dots represent individual data points. P values are indicated above the bars. (N.S., not significant P > 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; dark vs. light using two-tailed unpaired t-test).
Extended Data Fig. 7 RedPA-Cre activation with various red-light illumination conditions.
HEK 293T cells were transfected with plasmids encoding RedPA-Cre and the bioluminescence reporter without additional BV supplementation. Twenty-four hours after the transfection, the cells were illuminated with red light at different illumination durations and intensities. In the left panels showing the illumination conditions, gray corresponds to the dark condition and red with asterisk corresponds to the red light condition with denoted light intensities. Ratios of the mean bioluminescence intensity under the red light condition (red bar) to that under the dark condition (gray bar) are depicted above the bars. Bar data are shown as the mean ± s.d. from four biological replicates. Dots represent individual data points. P values are indicated above the bars. (****P < 0.0001; one-way ANOVA with multiple comparisons).
Extended Data Fig. 8 RedPA-Cre under dark, 800-nm illumination, and 660-nm illumination conditions.
DNA recombination activities of RedPA-Cre were compared among the dark, the 800-nm illumination and the 660-nm illumination conditions. Experimental conditions are the same as those in Fig. 5c, d except for that the 800-nm samples are incubated under 800-nm pulsed light (1 min ON and 4 min OFF) of 10 W m−2. Bar data are shown as the mean ± s.d. from four biological replicates. Dots represent individual data points. P values are indicated above the bars. (N.S., not significant P > 0.05; ****P < 0.0001; dark vs. 660 nm, dark vs. 800 nm and 800 nm vs. 660 nm using two-tailed unpaired t-test).
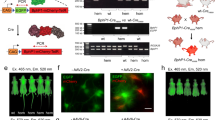
Extended Data Fig. 9 RedPA-Cre enables for DNA recombination reaction upon noninvasive red light illumination in living mice.
(a, b) ICR mice were transfected with a bioluminescent reporter plasmid together with pcDNA3.1 empty plasmid as a control (a) or with plasmid encoding RedPA-Cre of which configuration is NLS-DrBphP-CreC106-IRES-NLS-CreN104-Aff6_V18FΔN (b). Twenty-five hours after the transfection, bioluminescence images were obtained. No difference can be observed in the appearance between the mice maintained in the dark and the ones illuminated with red light at 660 nm for 16 h. The total bioluminescence intensities were shown in Fig. 5f (n = 3 mice per group).
Extended Data Fig. 10 In vivo gene activation by Red-CPTS upon noninvasive red light illumination.
(a, b) ICR mice were transfected with plasmids encoding Red-CPTS and luciferase reporter together with a plasmid encoding unrelated sgRNA as a negative control (a, Empty) or sgRNA targeting GAL4UAS (b, GAL4UAS). After the transfection, the mice were noninvasively illuminated at 660 nm or kept in the dark as shown in Supplementary Figure 28, and then bioluminescence imaging of the mice was performed. (c) Total bioluminescence intensities of the mice shown in a and b. Gray and red bars represent the mean ± s.d., and dots represent the total bioluminescence intensity of each mouse (n = 4 mice per group). (N.S. P > 0.05; ****P < 0.0001; using two-way ANOVA with multiple comparisons). (d) Red light-dependent endogenous gene activation by Red-CPTS with unrelated sgRNA as a negative control (Empty) or sgRNA targeting mouse ASCL1 (mASCL1) in vivo in living BALB/c mice. Data are represented as the relative mRNA level to the non-transfected negative control (n = 6 mice per group). Gray and red bars represent the mean ± s.d., and dots represent individual data points. No difference can be observed in the appearance between the mice maintained in the dark and the ones illuminated with red light at 660 nm for 16 h. P values are indicated above the bars. (N.S., not significant P > 0.05; ****P < 0.0001; using two-way ANOVA with multiple comparisons).
Supplementary information
Supplementary Information
Supplementary Figs. 1–20, Tables 1–4 and Notes.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Rights and permissions
About this article
Cite this article
Kuwasaki, Y., Suzuki, K., Yu, G. et al. A red light–responsive photoswitch for deep tissue optogenetics. Nat Biotechnol 40, 1672–1679 (2022). https://doi.org/10.1038/s41587-022-01351-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-022-01351-w
This article is cited by
-
A programmable protease-based protein secretion platform for therapeutic applications
Nature Chemical Biology (2024)
-
Integrated compact regulators of protein activity enable control of signaling pathways and genome-editing in vivo
Cell Discovery (2024)
-
Engineered poly(A)-surrogates for translational regulation and therapeutic biocomputation in mammalian cells
Cell Research (2024)
-
Programmable RNA base editing with photoactivatable CRISPR-Cas13
Nature Communications (2024)
-
Orthogonal LoxPsym sites allow multiplexed site-specific recombination in prokaryotic and eukaryotic hosts
Nature Communications (2024)