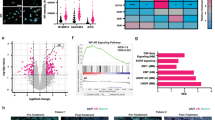
Abstract
Treatment with therapy targeting BRAF and MEK (BRAF/MEK) has revolutionized care in melanoma and other cancers; however, therapeutic resistance is common and innovative treatment strategies are needed1,2. Here we studied a group of patients with melanoma who were treated with neoadjuvant BRAF/MEK-targeted therapy (NCT02231775, n = 51) and observed significantly higher rates of major pathological response (MPR; ≤10% viable tumour at resection) and improved recurrence-free survival (RFS) in female versus male patients (MPR, 66% versus 14%, P = 0.001; RFS, 64% versus 32% at 2 years, P = 0.021). The findings were validated in several additional cohorts2,3,4 of patients with unresectable metastatic melanoma who were treated with BRAF- and/or MEK-targeted therapy (n = 664 patients in total), demonstrating improved progression-free survival and overall survival in female versus male patients in several of these studies. Studies in preclinical models demonstrated significantly impaired anti-tumour activity in male versus female mice after BRAF/MEK-targeted therapy (P = 0.006), with significantly higher expression of the androgen receptor in tumours of male and female BRAF/MEK-treated mice versus the control (P = 0.0006 and P = 0.0025). Pharmacological inhibition of androgen receptor signalling improved responses to BRAF/MEK-targeted therapy in male and female mice (P = 0.018 and P = 0.003), whereas induction of androgen receptor signalling (through testosterone administration) was associated with a significantly impaired response to BRAF/MEK-targeted therapy in male and female patients (P = 0.021 and P < 0.0001). Together, these results have important implications for therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The additional datasets generated during and/or analysed during the current study of clinical trial NCT02231775 are available at the European Genome–Phenome Archive (EGAS00001006196). Other datasets generated during and/or analysed during the current study are available from the corresponding authors on reasonable request.
Change history
10 January 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-05632-x
References
Long, G. V. et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 17, 1743–1754 (2016).
Robert, C. et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. New Engl. J. Med. 381, 626–636 (2019).
Flaherty, K. T. et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 367, 107–114 (2012).
Long, G. V. et al. Neoadjuvant dabrafenib combined with trametinib for resectable, stage IIIB-C, BRAFV600 mutation-positive melanoma (NeoCombi): a single-arm, open-label, single-centre, phase 2 trial. Lancet Oncol. 20, 961–971 (2019).
Leung, J. K. & Sadar, M. D. Non-genomic actions of the androgen receptor in prostate cancer. Front. Endocrinol. 8, 2 (2017).
Pinto, J. A. et al. Gender and outcomes in non-small cell lung cancer: an old prognostic variable comes back for targeted therapy and immunotherapy? ESMO Open 3, e000344 (2018).
Capone, I., Marchetti, P., Ascierto, P. A., Malorni, W. & Gabriele, L. Sexual dimorphism of immune responses: a new perspective in cancer immunotherapy. Front. Immunol. 9, 552 (2018).
Clocchiatti, A. et al. Androgen receptor functions as transcriptional repressor of cancer-associated fibroblast activation. J. Clin. Invest. 128, 5531–5548 (2018).
Pequeux, C. et al. Stromal estrogen receptor-alpha promotes tumor growth by normalizing an increased angiogenesis. Cancer Res. 72, 3010–3019 (2012).
Zhao, L. et al. Pharmacological activation of estrogen receptor beta augments innate immunity to suppress cancer metastasis. Proc. Natl Acad. Sci. USA 115, E3673–E3681 (2018).
Rothenberger, N. J., Somasundaram, A. & Stabile, L. P. The role of the estrogen pathway in the tumor microenvironment. Int. J. Mol. Sci. 19, 611 (2018).
Ribeiro, M. P. C., Santos, A. E. & Custodio, J. B. A. The activation of the G protein-coupled estrogen receptor (GPER) inhibits the proliferation of mouse melanoma K1735-M2 cells. Chem. Biol. Interact. 277, 176–184 (2017).
Qi, J. Therapy resistance by splicing: can the androgen receptor teach us about BRAF? Pigm. Cell Melanoma Res. 25, 293–294 (2012).
Wang, Y. et al. Androgen receptor promotes melanoma metastasis via altering the miRNA-539-3p/USP13/MITF/AXL signals. Oncogene 36, 1644–1654 (2017).
Natale, C. A. et al. Activation of G protein-coupled estrogen receptor signaling inhibits melanoma and improves response to immune checkpoint blockade. eLife 7, e31770 (2018).
Smalley, K. S. Why do women with melanoma do better than men? eLife 7, e33511 (2018).
Marzagalli, M. et al. Estrogen receptor beta in melanoma: from molecular insights to potential clinical utility. Front. Endocrinol. 7, 140 (2016).
Amaria, R. N. et al. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial. Lancet Oncol. 19, 181–193 (2018).
Stacchiotti, S. et al. High-grade soft-tissue sarcomas: tumor response assessment-pilot study to assess the correlation between radiologic and pathologic response by using RECIST and Choi criteria. Radiology 251, 447–456 (2009).
Eisenhauer, E. A. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2008).
Mendiratta, P. et al. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J. Clin. Oncol. 27, 2022–2029 (2009).
Schmidt, K. et al. The lncRNA SLNCR recruits the androgen receptor to EGR1-bound genes in melanoma and inhibits expression of tumor suppressor p21. Cell Rep 27, 2493–2507 (2019).
Ma, M. et al. Sustained androgen receptor signaling is a determinant of melanoma cell growth potential and tumorigenesis. J. Exp. Med. 218, e20201137 (2021).
Zhou, Z. X., Lane, M. V., Kemppainen, J. A., French, F. S. & Wilson, E. M. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol. Endocrinol. 9, 208–218 (1995).
Jin, H. J., Kim, J. & Yu, J. Androgen receptor genomic regulation. Transl. Androl. Urol. 2, 157–177 (2013).
Morvillo, V. et al. Atypical androgen receptor in the human melanoma cell line IIB-MEL-J. Pigm. Cell Res. 8, 135–141 (1995).
Aguirre-Portoles, C. et al. ZIP9 Is a druggable determinant of sex differences in melanoma. Cancer Res. 81, 5991–6003 (2021).
Liang, C. et al. TRIM36, a novel androgen-responsive gene, enhances anti-androgen efficacy against prostate cancer by inhibiting MAPK/ERK signaling pathways. Cell Death Dis. 9, 155 (2018).
Li, S. et al. Activation of MAPK signaling by CXCR7 leads to enzalutamide resistance in prostate cancer. Cancer Res. 79, 2580–2592 (2019).
Zhang, M. et al. Targeting AR-Beclin 1 complex-modulated growth factor signaling increases the antiandrogen enzalutamide sensitivity to better suppress the castration-resistant prostate cancer growth. Cancer Lett. 442, 483–490 (2019).
Wu, H. et al. Combination of sorafenib and enzalutamide as a potential new approach for the treatment of castration-resistant prostate cancer. Cancer Lett. 385, 108–116 (2017).
Kuser-Abali, G., Alptekin, A., Lewis, M., Garraway, I. P. & Cinar, B. YAP1 and AR interactions contribute to the switch from androgen-dependent to castration-resistant growth in prostate cancer. Nat. Commun. 6, 8126 (2015).
Lin, L. et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47, 250–256 (2015).
Rubin, J. B. et al. Sex differences in cancer mechanisms. Biol. Sex Differ. 11, 17 (2020).
Boese, A. C., Kim, S. C., Yin, K. J., Lee, J. P. & Hamblin, M. H. Sex differences in vascular physiology and pathophysiology: estrogen and androgen signaling in health and disease. Am. J. Physiol. Heart. Circ. Physiol. 313, H524–H545 (2017).
Guan, X. et al. Androgen receptor activity in T cells limits checkpoint blockade efficacy. Nature https://doi.org/10.1038/s41586-022-04522-6 (2022).
Balzano, S. et al. The effect of androgen blockade on pulsatile gonadotrophin release and LH response to naloxone. Clin. Endocrinol. 27, 491–499 (1987).
Kerrigan, J. R., Veldhuis, J. D. & Rogol, A. D. Androgen-receptor blockade enhances pulsatile luteinizing hormone production in late pubertal males: evidence for a hypothalamic site of physiologic androgen feedback action. Pediatr. Res. 35, 102–106 (1994).
Maughan, B. L. & Antonarakis, E. S. Enzalutamide in chemo-naive castration-resistant prostate cancer: effective for most but not for all. Asian J. Androl. 16, 807–808 (2014).
Dehm, S. M., Schmidt, L. J., Heemers, H. V., Vessella, R. L. & Tindall, D. J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 68, 5469–5477 (2008).
Arora, V. K. et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155, 1309–1322 (2013).
Cross, T. L., Kasahara, K. & Rey, F. E. Sexual dimorphism of cardiometabolic dysfunction: gut microbiome in the play? Mol. Metab. 15, 70–81 (2018).
Martin, A. M., Sun, E. W., Rogers, G. B. & Keating, D. J. The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front. Physiol. 10, 428 (2019).
Sudo, N. Microbiome, HPA axis and production of endocrine hormones in the gut. Adv. Exp. Med. Biol. 817, 177–194 (2014).
Gaballa, R. et al. Exosomes-mediated transfer of Itga2 promotes migration and invasion of prostate cancer cells by inducing epithelial-mesenchymal transition. Cancers 12, 2300 (2020).
Ricke, E. A. et al. Androgen hormone action in prostatic carcinogenesis: stromal androgen receptors mediate prostate cancer progression, malignant transformation and metastasis. Carcinogenesis 33, 1391–1398 (2012).
Scatena, C. et al. Androgen receptor expression inversely correlates with histological grade and N stage in ER+/PgRlow male breast cancer. Breast Cancer Res. Treat. 182, 55–65 (2020).
Wang, D. & Tindall, D. J. Androgen action during prostate carcinogenesis. Methods Mol. Biol. 776, 25–44 (2011).
Xia, N., Cui, J., Zhu, M., Xing, R. & Lu, Y. Androgen receptor variant 12 promotes migration and invasion by regulating MYLK in gastric cancer. J. Pathol. 248, 304–315 (2019).
McQuade, J. L. et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol. 19, 310–322 (2018).
Robert, C. et al. Five-year outcomes from a phase 3 METRIC study in patients with BRAF V600 E/K-mutant advanced or metastatic melanoma. Eur. J. Cancer 109, 61–69 (2019).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. Version 0.11.9. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2021).
Efstathiou, E. et al. Enzalutamide in combination with abiraterone acetate in bone metastatic castration-resistant prostate cancer patients. Eur. Urol. Oncol. 3, 119–127 (2020).
Maity, S. N. et al. Targeting of CYP17A1 lyase by VT-464 inhibits adrenal and intratumoral androgen biosynthesis and tumor growth of castration resistant prostate cancer. Sci. Rep. 6, 35354 (2016).
Acknowledgements
J.A.W. is supported by the NIH (1 R01 CA219896-01A1), the Melanoma Research Alliance (4022024), the American Association for Cancer Research Stand Up To Cancer (SU2C-AACR-IRG-19-17) and the MD Anderson Cancer Center’s Melanoma Moon Shots Program. J.L.M. is supported by the Melanoma Research Alliance, an American Society of Clinical Oncology and Conquer Cancer Foundation Career Development Award, the Elkins Foundation, the Seerave Foundation, the Rising Tide Foundation, the Mark Foundation, an MD Anderson Cancer Center (MDACC) Melanoma SPORE Developmental Research Program Award and the MD Anderson Physician Scientist Program, and acknowledges the Transdisciplinary Research in Energetics and Cancer Research Training Workshop R25CA203650 and the MDACC Center for Energy Balance in Cancer Prevention and Survivorship. A.P.C. is supported by the CPRIT Research Training Program at the MD Anderson Cancer Center (RP170067), The US Department of State, Bureau of Educational and Cultural Affairs, and the Fulbright Franco-Américaine Commission. M.C.A. is supported by a National Health and Medical Research Council of Australia CJ Martin Early Career Fellowship (1148680). B.H. was supported by the National Institutes of Health (T32 CA 009599) and the MD Anderson Cancer Center support grant (P30 CA016672). M.G.W. is supported by the National Institutes of Health (T32 CA 009599) and the MD Anderson Cancer Center support grant (P30 CA016672). R.G.W. is supported by the National Institutes of Health (T32 CA 009599) and the MD Anderson Cancer Center support grant (P30 CA016672). G.M. is supported by the National Institute of Health (1F32CA260769-01). M.A.D. is supported by the Dr Miriam and Sheldon G. Adelson Medical Research Foundation, the AIM at Melanoma Foundation, the NIH/NCI (1 P50 CA221703-02), Cancer Fighters of Houston, the Anne and John Mendelsohn Chair for Cancer Research, and philanthropic contributions to the Melanoma Moon Shots Program of MD Anderson. G.O. is supported by the National Institutes of Health (T32 CA 009599) and the MD Anderson Cancer Center support grant (P30 CA016672). The University of Texas MD Anderson Cancer Center Science Park Next-Generation Sequencing (NGS) Facility is supported by CPRIT Core Facility Support Awards (CPRIT RP170002).
Author information
Authors and Affiliations
Contributions
C.P.V., M.G.W., M.C.A. and J.A.W. formulated research goals and aims. C.P.V., M.G.W., M.C.A., T.P.H., J.L.M., J.R.M. and J.A.W. developed and designed methodology. M.G.W., M.C.A., L.W., H.Z., S.E.W., M.J.L., L.E.H., G.H. and Y.C. designed and executed pipelines and analytical code. C.P.V., M.G.W., R.G.W., M.C.A., H.Z., F.C., S.E.W., M.J.L., M.C., M.A.W.K., H.B., E.P. and L.P. conducted statistical analysis, built models and plotted results. C.P.V., J.R.M., J.L.M., M.T.T., M.G.W., M.T., C.L., J.R.D., M.M., R.G.W., Z.A.C., G.O., A.P.C., G.M., P.P., E.M.B., B.H., J.J.K., R.L.S., X.M. and Z.K. designed and conducted immunological assays and preclinical studies, generated and annotated metadata and created cell lines. J.L.M., P.A.F., M.A.D., R.N.A., H.A.T., S.P., P.H., J.E.L., J.E.G., A.L., E.Z.K., M.I.R., A.J.L., K.W., C.W.H. and G.V.L. provided clinical samples. J.J.K., S.J., N.F., G.G., E.L., Q.L. and M.S.B. performed in vitro studies. C.P.V. and J.R.M. were scientific drivers, and planned and oversaw all mouse experiments and studies. M.P. generated of AR-knockout cell lines. M.G.W., C.P.V., M.C.A., L.F., J.R.D., J.L.M., J.A.W., R.G.W., M.C., S.A., M.S.B., M.S.Y. and S.N.W. prepared the manuscript. A.E.M., T.P.H., J.R.M. and J.A.W. provided scientific oversight. J.A.W. supervised and oversaw all aspects of study including design, conduct and analyses. All of the authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.A.W. is listed as an inventor on a US patent application (PCT/US17/53.717); reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, MedImmune and Bristol-Myers Squibb (BMS); serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline (GSK), BMS, Merck, Biothera Pharmaceuticals and Micronoma; and holds stock options from Micronoma. J.L.M. serves as a consultant for Merck and is on the advisory board for BMS. Z.A.C. is a current employee of and owns stock in AstraZeneca. M.C.A. reports advisory board participation and honoraria from MSD Australia outside the current work; received research funds to their institution from MSD Australia and BMS Australia outside the current work; and is a named co-inventor on patent applications relating to methods and compositions for treating cancer (WO2020106983A1) unrelated to the current work, and methods for improving sex-dimorphic responses to targeted therapy in melanoma (US20200164034A1) related to the current work. R.N.A. receives research support and advisory boards for Novartis, BMS, Iovance and research support from Genentech. C.L. serves as a consultant/advisory board for Merck, Sharp & Dohme, Exelixis, Bayer and Amgen; reports honoraria from Merck, Sharp & Dohme, Bayer and Amgen; and clinical grants from Janssen, ORIC Pharmaceuticals, Novartis and Aragon Pharmaceuticals. M.A.D. is an advisory board member/consultant for BMS, Novartis, Array, Roche/Genentech, GSK, Sanofi-Aventis and ABM Therapeutics; principal investigator of a research grant to their institution from Astrazeneca, Roche/Genentech, GSK, Myriad, Oncothyreon, Sanofi-Aventis and ABM Therapeutics. P.A.F. serves on the scientific advisory board for Scorpion Therapeutics. A.E.M. has a sponsored research agreement with MedImmune/AstraZeneca. S.N.W. serves as a consultant for Agenus, AstraZeneca, Caris, Clovis Oncology, Eisai, EQRX, GSK/Tesaro, ImmunoGen, Lilly, Merck, Pfizer, Mersana, Roche/Genentech and Zentalis; she receives research support to her institution from AstraZeneca, Bayer, Bio-Path, Clovis Oncology, Cotinga Pharmaceuticals, GSK/Tesaro, Mereo, Novartis, OncXerna, Roche/Genentech and Zentalis. A.J.L. has served on advisory boards and/or consulted for AbbVie, AstraZeneca, Bayer, BMS, Deciphera Pharmaceuticals, Foghorn Therapeutics, GSK, Invitae, Illumina, Iterion Therapeutics, Merck, Novartis, Nucleai, Paige.AI, Pfizer, Roche/Genentech and ThermoFisher. G.V.L. is a consultant advisor for Agenus, Amgen, Array Biopharma, Boehringer Ingelheim International, Bristol Myers Squibb, Evaxion Biotech A/S, Hexal (Sandoz Company), Highlight Therapeutics S.L., Merck Sharpe & Dohme (Australia) Pty Limited, Novartis Pharma, OncoSec Medical Australia, Pierre Fabre, Provectus, Qbiotics Group Limited, Regeneron Pharmaceuticals. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Caroline Robert, Joshua Rubin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Expanded analysis of clinical cohorts.
A) Rates of major pathologic response (MPR) based on menopausal status of female patients within the neoadjuvant cohort (p = 0.79) by logistic regression. B) Waterfall plot of percent change in tumour size by cross-sectional imaging in the neoadjuvant cohort (p = 0.54). C) Waterfall plot of percent change in tumour size by cross-sectional imaging of the pooled neoadjuvant and metastatic cohorts (p = 0.02).
Extended Data Fig. 2 Analysis of published cohorts of BRAF/MEK inhibited patients.
A) Schema of metastatic BRAF inhibitor monotherapy clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. B) Progression free survival by sex in the metastatic BRAF inhibitor monotherapy clinical cohort (n = 212, hazard ratio 1.2 CI [0.88-1.6] p = 0.241, by Kaplan-Meier method). C) Overall survival by sex in the metastatic BRAF inhibitor monotherapy clinical cohort (n = 212, hazard ratio 1.32 CI [0.95-1.82] p = 0.095, by Kaplan-Meier method). D) Schema of metastatic MEK inhibitor monotherapy clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. E) Progression free survival by sex in the metastatic MEK inhibitor monotherapy clinical cohort (n = 206, hazard ratio 1.35 CI [1.01-1.81] p = 0.043, by Kaplan-Meier method). F) Overall survival by sex in the metastatic MEK inhibitor monotherapy clinical cohort (n = 206, hazard ratio 1.61 CI [1.19-2.18] p = 0.002, by Kaplan-Meier method). G) Schema of second neoadjuvant clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. H) Major pathologic response defined as ≤ 10% viable tumour in females versus males (p = 0.92, by Chi-squared). I) Recurrence free survival by sex in the second neoadjuvant cohort (n = 40, hazard ratio 1.16, 95% CI [0.36–2.07], p = 0.733, by Kaplan-Meier method). J) Kaplan Meier survival curves of overall survival by sex of melanoma patients abstracted with The Cancer Genome Atlas (p = 0.373 by univariate analysis, p = 0.369 controlling for age at diagnosis and stage). K) Paired clinical samples of circulating testosterone prior to initiation and after treatment with BRAF/MEK targeted therapy (p = 0.9453 and 0.2036 respectively by two-sided Student’s t-test). L) AR staining pre-treatment in males (blue) and females (pink) by MPR (p = 0.72, by two-sided unpaired t-test).
Extended Data Fig. 3 Transcriptomic analysis of clinical specimens.
A) Heatmap of AR signature genes of on-treatment specimens demonstrating upregulation of AR signature pathways. B) Volcano plot of differentially expressed genes after whole transcriptomic analysis of on-treatment with BRAF/MEK inhibition clinical specimens. Vertical dotted blue lines represent log foldchange greater than 2. Horizontal dotted blue line represents q < 0.05. Genes with log fold change greater than 5 and q < 0.05 are labelled. C) Heatmap of AR signature genes of pre-treatment specimens of AR signature pathways. D) Volcano plot of differentially expressed genes after whole transcriptomic analysis of pre-treatment with BRAF/MEK inhibition clinical specimens. E) Androgen signalling score comparing clinical samples achieving an MPR and those not achieving an MPR for those on treatment (ON, <MPR n = 2, MPR n = 4), at the time of resection (SURG, <MPR n = 9, MPR = 7; p = 0.066), aggregate of those on treatment and surgically resected (ONandSURG, <MPR = 11, MPR = 11; p = 0.011), and samples collected prior to treatment (PRE, <MPR n = 4, MPR n = 5; p = 0.95); groups were compared using a two-sided Student’s t test. Box plot represents the median bar with the box bounding interquartile range (IQR) and whiskers to the most extreme point within 1.5 x IQR.
Extended Data Fig. 4 Murine model of melanoma validates a sexually dimorphic response and suggests AR activity as a mechanism of resistance.
A-B) Percent change in tumour volume for male and female C57BL/6 mice implanted subcutaneously with BP cells that were treated with Vehicle or BRAF/MEKi (n = 10 mice per group; A – mice aged 9 weeks, B - mice aged 12 weeks). Results from the second and third repeats of this experiment are shown in A and B, respectively (p = 0.039 and p = 0.45). C) Percent change in tumour volume in BP injected C57BL/6 male mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation through androgen blockade with enzalutamide or castration (mice aged 14 weeks). All tumour growth curves were compared by ANOVA with multiple comparisons (n = 10/group except BRAF/MEKi + castration where n = 9; p = 0.003 BRAF/MEKi vs BRAF/MEKi + Enzalutamide; p = 0.031 BRAF/MEKi vs BRAF/MEKi + Castration). D-E) Percent change in tumour volume for AR-KO BP tumours in female (D) (p = 0.99) and male (E) (p = 0.98) CD-1 mice treated with vehicle or BRAF/MEKi in combination with either testosterone or enzalutamide, respectively (n = 10 mice/group; aged 11 weeks). All tumour growth represented as mean + SEM and p-values were calculated using ANOVA with multiple comparisons.
Extended Data Fig. 5 Mechanistic and Validations Murine Studies Tumour Volume Curves.
A) Tumour volumes for male and female C57BL/6 mice implanted subcutaneously with YUMMER 1.7 cells treated with Vehicle or BRAF/MEK inhibition (p = 0.06 between male and female BRAF/MEKi; 30 mpk dabrafenib and 1 mpk trametinib, PO, QD). n = 10 mice per group, aged 12-13 weeks. B-D) Percent change in tumour volume for male and female C57BL/6 mice implanted subcutaneously with BP (BRAFV600E, PTEN−/−) cells (p = 0.003, 0.26, and 0.93) between male and female BRAF/MEKi. Mice were treated as in A. n = 10 mice per group; aged 12-13 weeks. The experiment was performed in triplicate with each experiment represented above. E) Tumour volumes for male and female CD-1 nude mice implanted and treated as in A (p = 0.01 between male and female BRAF/MEKi). n = 10 mice per group; aged 11 weeks. F) Tumour volumes of male and female CD-1 mice with AR-KO BP tumours treated with vehicle or BRAF/MEKi (p = 0.317 between male and female BRAF/MEKi); n = 10/group; aged 11 weeks. All tumour growth represented as mean + SEM and p-values were calculated using ANOVA with multiple comparisons.
Extended Data Fig. 6 AR gene and protein expression analysis of preclinical models and serum testosterone measurements of clinical specimens.
A) AR gene expression of murine BP tumours from male and female mice treated with either Vehicle or BRAFi/MEKi (female Vehicle vs Female BRAF/MEKi treated, p = 0.005, male Vehicle vs male BRAF/MEKi treated, p = 0.16) p-values were calculated using two-sided Student’s t-test B) AR immunofluorescence staining of samples from female and male mice treated with Vehicle, BRAF/MEKi, BRAF/MEKI + Testosterone (females, vehicle vs BRAF/MEKi, p = 0.005) and BRAF/MEKi + enzalutamide (males, vehicle vs BRAF/MEKi, p = 0.16). p-values were calculated by two-sided Student’s t-test. C) Plasma testosterone levels for male mice across treatment groups. Decreased plasma testosterone was noted in both the castration group as compared to Vehicle (p = 0.039) as well as the BRAF/MEKi + castration group as compared to the BRAF/MEKi group (p = 0.017). p-values were calculated using a Kruskal Wallis test. D) Plasma testosterone levels for female mice across treatment groups. Increased plasma testosterone was noted in the Vehicle + testosterone group as compared to the BRAF/MEKi group (p = 0.007) and vehicle group (p = 0.007). Similarly increased testosterone was noted in the BRAF/MEKi + testosterone group as compared to the vehicle group (p = 0.009) and BRAF/MEKi group (p = 0.009). No other associations were significant p < 0.05 by Kruskal Wallis test. E) AR immunofluorescence staining of AR-KO BP tumour samples from female and male mice treated with either Vehicle or BRAFi/MEKi. p-values were calculated by Student’s t-test. Histograms in A and B represent mean + SD whereas C and D represent mean + SEM.
Extended Data Fig. 7 Modulation of AR activity is associated with differential response to BRAF/MEK inhibition.
A) Percent change in tumour volume in BP injected C57BL/6 female mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation with either estradiol or estradiol and oophorectomy (n = 10/group; mice aged 14 weeks). No significant effects exist within either cohort as calculated by ANOVA. B) Percent change in tumour volume for female C57BL/6 mice implanted subcutaneously with YUMMER 1.7 cells treated with BRAF/MEKi or BRAF/MEKi + testosterone (n = 10/group; aged 13 weeks, p = 0.0003 between BRAF/MEKi and BRAF/MEKi + testosterone). C) Percent change in tumour volume in YUMMER1.7 injected into C57BL/6 male mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation with enzalutamide (n = 10/group; mice aged 14 weeks, p = 0.031). D) Heatmap of differentially expressed androgen responsive genes between high versus low testosterone groups q < 0.05. Groups by androgen staining levels, sex, and treatment. E) Percent change in BP tumour volume in male C57BL/6 mice treated with vehicle in the presence or absence of BRAF/MEKi with physical castration or BRAF/MEKi with castration and exogenous testosterone (n = 10/group; aged 14 weeks, p = 0.01 for BRAF/MEKi + castration versus BRAF/MEKi, p = 0.04 for BRAF/MEKi versus BRAF/MEKi + castration + testosterone, and p = 0.0004 for BRAF/MEKi + castration vs BRAF/MEKi + castration + testosterone). F) Quantification of the percent of AR+ nuclei by immunofluorescence in BP tumours from male mice treated with vehicle (n = 7), BRAF/MEKi (n = 5), or BRAF/MEKi + enzalutamide (n = 7) (p = 0.003 between BRAF/MEKi and BRAF/MEKi + enzalutamide). p-values were calculated using two-sided Student’s t-test. G) Quantification of the percent of AR+ nuclei by immunofluorescence in BP tumours from female mice treated with vehicle (n = 7), BRAF/MEKi (n = 7), BRAF/MEKi + testosterone (n = 5) (p = 0.006 between vehicle and BRAF/MEKi and p = 0.003 between BRAF/MEKi and BRAF/MEKi + testosterone in female mice). p-values were calculated using two-sided Student’s t-test. Tumour growth curves in panels A-C and E represent mean + SEM. Histograms in panels F and G represent mean + SEM. All tumour growth curves were compared by ANOVA with multiple comparisons.
Extended Data Fig. 8 Interventional Murine Studies Tumour Volumes.
A) Tumour volumes curves of BP tumour growth in male mice treated with vehicle (n = 10), BRAF/MEKi alone (n = 9) or BRAF/MEKi in combination with testosterone (n = 9) (p = 0.12). B) Tumour volumes curves of BP tumour growth in female mice treated with vehicle, BRAF/MEKi alone or BRAF/MEKi in combination with testosterone (p = 0.003). (n = 10 mice/group). C) Tumour volume curves of BP tumour growth in male mice treated with BRAF/MEKi alone or BRAF/MEKi in combination with AR blockade (p = 0.02) or AR blockade with testosterone (p = 0.07) (n = 10 mice/group). D) Tumour volume curves of BP tumour growth in female mice treated with BRAF/MEKi alone or BRAF/MEKi in combination with AR blockade (p = 0.002) or AR blockade with testosterone (p = 0.34) (n = 10 mice/group). E) Tumour volume curves of BP AR KO tumour growth in CD-1 nude female mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation (mice aged 14 weeks). BRAF/MEKi vs BRAF/MEKi + testosterone, p = 0.99; (n = 10 mice/group). F) Tumour volume curves of AR KO BP injected CD-1 nude male mice treated with vehicle in the presence or absence of BRAF/MEKi or BRAF/MEKi + enzalutamide (p = 0.98) (n = 10 mice/group). G) Tumour volume curves of YUMMER 1.7 tumour growth in male mice treated with vehicle, BRAF/MEKi or BRAF/MEKi + AR blockade (p = 0.12) (n = 10 mice/group) H) Tumour volume curves of BP tumour growth in C57BL/6 male mice treated with BRAF/MEKi, BRAF/MEKi + castration (p = 0.01) or BRAF/MEKi + castration + testosterone (p = 0.0005) (n = 10 mice/group). I) Change in tumour volume in BP injected C57BL/6 male mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation through either androgen blockade, androgen blockade with testosterone, estradiol, or castration (mice aged 14 weeks) (n = 10 mice/group). J) Tumour volume curves of YUMMER 1.7 tumour growth in female mice treated with vehicle, BRAF/MEKi or BRAF/MEKi + testosterone (p = 0.0005) (n = 10 mice/group). K) Change in tumour volume in BP injected C57BL/6 female mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation with either estradiol or estradiol and oophorectomy (n = 10 mice/group; mice aged 14 weeks). L) Change in tumour volume of BP tumours injected into CD-1 nude male mice and treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with AR blockade (n = 10 mice/group, mice aged 11 weeks, BRAF/MEKi vs BRAF/MEKi + AR blockade p = 0.92). M) Change in tumour volume of BP tumours injected into CD-1 nude female mice and treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with testosterone (n = 10 mice/group, mice aged 11 weeks, BRAF/MEKi vs BRAF/MEKi + testosterone p = 0.03). All tumour growth curves represent mean + SEM and were compared by ANOVA with multiple comparisons.
Extended Data Fig. 9 Effect of BRAF/MEKi on pERK, ZIP9/YAP1 associated transcripts and YAP1 associated transcripts in BP tumours.
A) Staining and quantification of phosphor-ERK in BP tumours of female and male mice on treated with vehicles, BRAF/MEK inhibition, or BRAF/MEK inhibition + testosterone. Histogram represent mean + SD. Differences were calculated using one-way ANOVA (n = 5/group). B) Staining and quantification of ZIP9/YAP1 associated transcripts in BP tumours of female and male mice on treatment with vehicles (n = 6), BRAF/MEK inhibition (n = 4), or BRAF/MEK inhibition + testosterone (n = 8). Histogram represent mean + SD. Differences were calculated using one-way ANOVA. C) Staining and quantification of YAP1 associated transcripts in BP tumours of female and male mice on treatment with vehicles (n = 6 females, 4 males) or BRAF/MEK inhibition (n = 4 females, 3 males. Histogram represent mean + SD. Differences were calculated using one-way ANOVA.
Extended Data Fig. 10 Non-targeting control line in BRAF/PTEN mice.
A) Volume of BP tumours established from the BP cell line engineered to express a non-targeting CRISPR control for male CD-1 nude mice treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with AR blockade (n = 10 mice/group) B) Volume of BP tumours established from the BP cell line engineered to express a non-targeting CRISPR control for female CD-1 nude mice treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with testosterone (n = 10 mice/group, p = 0.01).). All tumour growth curves represent mean + SEM and were compared by ANOVA with multiple comparisons.
Supplementary information
Supplementary Table 1
MD Anderson clinical cohort demographic and clinical data. Metadata for 51 patients who received neoadjuvant BRAF/MEKi targeted therapy for melanoma and 80 patients with metastatic melanoma who received definitive BRAF/MEKi targeted therapy. MPR+, major pathological response; MPR−, no major pathologic response; OR, odds ratio; ECOG, Eastern Cooperative Oncology Group; BMI, body mass index; LDH, lactate dehydrogenase. MPR is defined as less than 10% viable tumour on pathological examination. Clinical benefit is defined as stable disease for >6 months, partial response or complete response on the basis of RECIST 1.1 criteria.
Supplementary Table 2
Detailed clinical characteristics of MD Anderson metastatic melanoma cohort with RECIST 1.1 evaluable data. Metadata of patients with metastatic melanoma treated at MD Anderson included in the study with evaluable RECIST 1.1 data. IQR, interquartile range. Clinical benefit is defined as stable disease for >6 months, partial response or complete response on the basis of RECIST 1.1 criteria.
Supplementary Table 3
Percentage of AR-positive nuclei by sex, MPR and RECIST response. Percentage of AR-positive nuclei on the basis of immunofluorescence staining with associated sex, pathological responses and RECIST responses.
Supplementary Table 4
Differentially expressed genes of on-treatment clinical samples. Clinical samples underwent bulk RNA-seq with the following genes being differentially expressed in on-treatment samples that achieved a MPR to those that did not achieve a MPR.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vellano, C.P., White, M.G., Andrews, M.C. et al. Androgen receptor blockade promotes response to BRAF/MEK-targeted therapy. Nature 606, 797–803 (2022). https://doi.org/10.1038/s41586-022-04833-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04833-8
This article is cited by
-
Sexual dimorphism in melanocyte stem cell behavior reveals combinational therapeutic strategies for cutaneous repigmentation
Nature Communications (2024)
-
Sex differences orchestrated by androgens at single-cell resolution
Nature (2024)
-
Outcome differences by sex in oncology clinical trials
Nature Communications (2024)
-
Androgen drives melanoma invasiveness and metastatic spread by inducing tumorigenic fucosylation
Nature Communications (2024)
-
Dyskerin and telomerase RNA component are sex-differentially associated with outcomes and Sunitinib response in patients with clear cell renal cell carcinoma
Biology of Sex Differences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.