Abstract
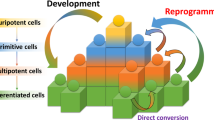
The reprogramming of somatic cells with defined factors, which converts cells from one lineage into cells of another, has greatly reshaped our traditional views on cell identity and cell fate determination. Direct reprogramming (also known as transdifferentiation) refers to cell fate conversion without transitioning through an intermediary pluripotent state. Given that the number of cell types that can be generated by direct reprogramming is rapidly increasing, it has become a promising strategy to produce functional cells for therapeutic purposes. This Review discusses the evolution of direct reprogramming from a transcription factor-based method to a small-molecule-driven approach, the recent progress in enhancing reprogrammed cell maturation, and the challenges associated with in vivo direct reprogramming for translational applications. It also describes our current understanding of the molecular mechanisms underlying direct reprogramming, including the role of transcription factors, epigenetic modifications, non-coding RNAs, and the function of metabolic reprogramming, and highlights novel insights gained from single-cell omics studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Waddington, C. H. The Strategy of the Genes. A Discussion of Some Aspects of Theoretical Biology. With an Appendix by H. Kacser (George Allen & Unwin, Ltd., 1957).
Davis, R. L., Weintraub, H. & Lassar, A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). Davis et al. demonstrated, for the first time, that the overexpression of one transcription factor could rewrite cell fate in vitro.
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Yamanaka, S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10, 678–684 (2012).
Buganim, Y., Faddah, D. A. & Jaenisch, R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 14, 427–439 (2013).
Takahashi, K. & Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17, 183–193 (2016).
Smith, Z. D., Sindhu, C. & Meissner, A. Molecular features of cellular reprogramming and development. Nat. Rev. Mol. Cell Biol. 17, 139–154 (2016).
Srivastava, D. & DeWitt, N. In vivo cellular reprogramming: the next generation. Cell 166, 1386–1396 (2016).
Jorstad, N. L. et al. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature 548, 103–107 (2017).
Li, H. & Chen, G. In vivo reprogramming for CNS repair: regenerating neurons endogenous glial cells. Neuron 91, 728–738 (2016).
Qian, L. et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598 (2012). Qian et al. demonstrated the feasibility of using in vivo direct reprogramming for heart repair.
Cahan, P. et al. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 (2014).
Stone, N. R. et al. Context-specific transcription factor functions regulate epigenomic and transcriptional dynamics during cardiac reprogramming. Cell Stem Cell 25, 87–102.e9 (2019).
Kulessa, H., Frampton, J. & Graf, T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9, 1250–1262 (1995).
Xie, H., Ye, M., Feng, R. & Graf, T. Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676 (2004).
Laiosa, C. V., Stadtfeld, M., Xie, H., de Andres-Aguayo, L. & Graf, T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25, 731–744 (2006).
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J. & Melton, D. A. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455, 627–632 (2008).
Ieda, M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010). Idea et al. identified reprogramming factors that could reprogramme mouse cardiac fibroblasts to cardiomyocyte-like cells in vitro.
Song, K. et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604 (2012).
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010). Vierbuchen et al. identified a combination of three factors to directly convert mouse fibroblasts into functional neurons in vitro.
Sekiya, S. & Suzuki, A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 (2011).
Huang, P. et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 (2011).
Yoo, A. S. et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 (2011).
Jayawardena, T. M. et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110, 1465–1473 (2012).
Jayawardena, T. M. et al. MicroRNA induced cardiac reprogramming in vivo. Circ. Res. 116, 418–424 (2015).
Xu, J., Du, Y. & Deng, H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16, 119–134 (2015).
Takahashi, J. Strategies for bringing stem cell-derived dopamine neurons to the clinic: the Kyoto trial. in Progress in Brain Research 230, 213–226 (Elsevier B.V., 2017).
Barker, R. A., Parmar, M., Studer, L. & Takahashi, J. Human trials of stem cell-derived dopamine neurons for Parkinson’s disease: dawn of a new era. Cell Stem Cell 21, 569–573 (2017).
Zarbin, M., Sugino, I. & Townes-Anderson, E. Concise review: update on retinal pigment epithelium transplantation for age-related macular degeneration. Stem Cell Transl. Med. 8, 466–477 (2019).
Blau, H. M. & Daley, G. Q. Stem cells in the treatment of disease. N. Engl. J. Med. 380, 1748–1760 (2019).
Li, H. & Chen, G. In vivo reprogramming for CNS repair: regenerating neurons from endogenous glial cells. Neuron 91, 728–738 (2016).
Jayawardena, T. M. et al. MicroRNA induced cardiac reprogramming in vivo evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 116, 418–424 (2014).
Niu, W. et al. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175 (2013).
Grande, A. et al. Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat. Commun. 4, 2373 (2013).
Guo, Z. et al. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14, 188–202 (2014).
Torper, O. et al. Generation of induced neurons via direct conversion in vivo. Proc. Natl Acad. Sci. USA 110, 7038–7043 (2013).
Heinrich, C. et al. Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Rep. 3, 1000–1014 (2014).
Su, Z., Niu, W., Liu, M. L., Zou, Y. & Zhang, C. L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 5, 3338 (2014).
Song, G. et al. Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell 18, 797–808 (2016).
Yao, K. et al. Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 560, 484–488 (2018).
Fu, J. D. & Srivastava, D. Direct reprogramming of fibroblasts into cardiomyocytes for cardiac regenerative medicine. Circ. J. 79, 245–254 (2015).
Gascón, S., Masserdotti, G., Russo, G. L. & Götz, M. Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell 21, 18–34 (2017).
Li, W. et al. Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells. Nat. Biotechnol. 32, 1223–1230 (2014).
Thorel, F. et al. Conversion of adult pancreatic α-cells to B-cells after extreme B-cell loss. Nature 464, 1149–1154 (2010).
Humeres, C. & Frangogiannis, N. G. Fibroblasts in the infarcted, remodeling, and failing heart. JACC Basic. Transl. Sci. 4, 449–467 (2019).
Jin, Y. et al. Three-dimensional brain-like microenvironments facilitate the direct reprogramming of fibroblasts into therapeutic neurons. Nat. Biomed. Eng. 2, 522–539 (2018).
Li, Y. et al. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci. Rep. 6, 38815 (2016).
Magnusson, J. P. et al. A latent neurogenic program in astrocytes regulated by Notch signaling in the mouse. Science 346, 237–241 (2014).
Hu, X. et al. Region-restrict astrocytes exhibit heterogeneous susceptibility to neuronal reprogramming. Stem Cell Rep. 12, 290–304 (2019).
Buffo, A. et al. Origin and progeny of reactive gliosis: a source of multipotent cells in the injured brain. Proc. Natl Acad. Sci. USA 105, 3581–3586 (2008).
Kang, S. H., Fukaya, M., Yang, J. K., Rothstein, J. D. & Bergles, D. E. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681 (2010).
De La Rossa, A. et al. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat. Neurosci. 16, 193–200 (2013).
Rouaux, C. & Arlotta, P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 15, 214–221 (2013).
Xiao, X. et al. Endogenous reprogramming of alpha cells into beta cells, induced by viral gene therapy, reverses autoimmune diabetes. Cell Stem Cell 22, 78–90.e4 (2018).
Collombat, P. et al. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into α and subsequently β cells. Cell 138, 449–462 (2009).
Bramswig, N. C. et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Invest. 123, 1275–1284 (2013).
Liu, Y. et al. Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J. Neurosci. 35, 9336–9355 (2015).
Colasante, G. et al. Rapid conversion of fibroblasts into functional forebrain GABAergic interneurons by direct genetic reprogramming. Cell Stem Cell 17, 719–734 (2015).
Li, W. et al. In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. eLife 3, 1846 (2014).
Nam, Y. J. et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 141, 4267–4278 (2014).
Miyamoto, K. et al. Direct in vivo reprogramming with sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell 22, 91–103.e5 (2018).
Lee, K. et al. Peptide-enhanced mRNA transfection in cultured mouse cardiac fibroblasts and direct reprogramming towards cardiomyocyte-like cells. Int. J. Nanomed. 10, 1841–1854 (2015).
Chakraborty, S. et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep. 3, 940–947 (2014).
Liu, Y. et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 23, 758–771.e8 (2018).
Chang, Y. et al. Efficient in vivo direct conversion of fibroblasts into cardiomyocytes using a nanoparticle-based gene carrier. Biomaterials 192, 500–509 (2019).
Fu, Y. et al. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 25, 1013–1024 (2015).
Park, G. et al. Conversion of mouse fibroblasts into cardiomyocyte-like cells using small molecule treatments. Biomaterials 54, 201–212 (2015).
Cao, N. et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216–1220 (2016).
Wang, Y. et al. Chemical conversion of mouse fibroblasts into functional dopaminergic neurons. Exp. Cell Res. 347, 283–292 (2016).
Dai, P., Harada, Y. & Takamatsu, T. Highly efficient direct conversion of human fibroblasts to neuronal cells by chemical compounds. J. Clin. Biochem. Nutr. 56, 166–170 (2015).
Li, X. et al. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203 (2015).
Zhang, L. et al. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 17, 735–747 (2015).
Yu, C., Liu, K., Tang, S. & Ding, S. Chemical approaches to cell reprogramming. Curr. Opin. Genet. Dev. 28, 50–56 (2014).
Li, J. & Mooney, D. J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 1, 16071 (2016).
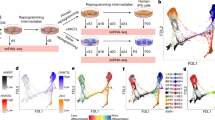
Liu, Z. et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 551, 100–104 (2017). Liu et al. constructed the routes of iCM formation using single-cell RNA-seq and discovered the critical role of splicing factor PTBP1 during murine cardiac reprogramming.
Zhou, Y. et al. Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell 25, 149–164.e9 (2019). Zhou et al. revealed an early decision point at which cells either continue reprogramming or regress toward the original fibroblast state during human cardiac reprogramming.
Treutlein, B. et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395 (2016). Treutlein et al. described the existence of an alternative reprogramming route in murine neuronal reprogramming using single-cell RNA-seq.
Richards, D. J. et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 4, 446–462 (2020).
Noor, N. et al. 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. 6, 1900344 (2019).
Zhou, Y. et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 18, 382–395 (2016).
Qian, H. et al. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 582, 550–556 (2020).
Trokovic, R., Weltner, J., Noisa, P., Raivio, T. & Otonkoski, T. Combined negative effect of donor age and time in culture on the reprogramming efficiency into induced pluripotent stem cells. Stem Cell Res. 15, 254–262 (2015).
Lo Sardo, V. et al. Influence of donor age on induced pluripotent stem cells. Nat. Biotechnol. 35, 69–74 (2017).
Soufi, A. et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015).
Iwafuchi-Doi, M. & Zaret, K. S. Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014).
Cirillo, L. A. et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002).
Wapinski, O. L. et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635 (2013). Wapinski et al. describe the role of Ascl1 as an ‘on-target’ Pioneer factor and its association to the ‘trivalent’ chromatin state in murine neuronal reprogramming.
Soufi, A., Donahue, G. & Zaret, K. S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004 (2012).
Chanda, S. et al. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 3, 282–296 (2014).
Hashimoto, H. et al. Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell 25, 69–86.e5 (2019).
Zhou, H., Dickson, M. E., Kim, M. S., Bassel-Duby, R. & Olson, E. N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl Acad. Sci. USA 112, 11864–11869 (2015).
Wang, L. et al. Improved generation of induced cardiomyocytes using a polycistronic construct expressing optimal ratio of Gata4, Mef2c and Tbx5. J. Vis. Exp. 105, 53426 (2015).
Wang, L. et al. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 116, 237–244 (2015).
Mathison, M. et al. ‘Triplet’ polycistronic vectors encoding Gata4, Mef2c, and Tbx5 enhances postinfarct ventricular functional improvement compared with singlet vectors. J. Thorac. Cardiovasc. Surg. 148, 1656–1664.e2 (2014).
Horisawa, K. et al. The dynamics of transcriptional activation by hepatic reprogramming factors. Mol. Cell 79, 660–676.e8 (2020).
Wapinski, O. L. et al. Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20, 3236–3247 (2017).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Liu, Z. et al. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 16, 507–518 (2016).
Kim, J. et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl Acad. Sci. USA 108, 7838–7843 (2011).
Barbagiovanni, G. et al. KMT2B is selectively required for neuronal transdifferentiation, and its loss exposes dystonia candidate genes. Cell Rep. 25, 988–1001 (2018).
Benayoun, B. A. et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158, 673–688 (2014).
Ezhkova, E. et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 25, 485–498 (2011).
Dal-Pra, S., Hodgkinson, C. P., Mirotsou, M., Kirste, I. & Dzau, V. J. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by MIR Combo. Circulation Res. 120, 1403–1413 (2017).
Becker, J. S., Nicetto, D. & Zaret, K. S. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 32, 29–41 (2016).
Huang, P. et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384 (2014).
Becker, J. S. et al. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell 68, 1023–1037.e15 (2017).
Hirai, H. & Kikyo, N. Inhibitors of suppressive histone modification promote direct reprogramming of fibroblasts to cardiomyocyte-like cells. Cardiovasc. Res. 102, 188–190 (2014).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Rhee, C. et al. Mechanisms of transcription factor-mediated direct reprogramming of mouse embryonic stem cells to trophoblast stem-like cells. Nucleic Acids Res. 45, 10103–10114 (2017).
Cao, J. & Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26 (2012).
Vastenhouw, N. L. & Schier, A. F. Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 24, 374–386 (2012).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Santoro, S. W. & Dulac, C. Histone variants and cellular plasticity. Trends Genet. 31, 516–527 (2015).
Henikoff, S. & Smith, M. M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7, a019364 (2015).
Gaspar-Maia, A. et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 4, 1565 (2013).
Dong, F. et al. Dynamic changes in occupancy of histone variant H2a.Z during induced somatic cell reprogramming. Stem Cells Int. 2016, 3162363 (2016).
Fang, H. T. et al. Global H3.3 dynamic deposition defines its bimodal role in cell fate transition. Nat. Commun. 9, 1537 (2018).
Cheng, H. et al. Reprogramming mouse fibroblasts into engraftable myeloerythroid and lymphoid progenitors. Nat. Commun. 7, 13396 (2016).
Luo, C. et al. Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. eLife 8, e40197 (2019).
Luginbühl, J., Sivaraman, D. M. & Shin, J. W. The essentiality of non-coding RNAs in cell reprogramming. Noncoding RNA Res. 2, 74–82 (2017).
Muraoka, N. et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 33, 1565–1581 (2014).
Xue, Y. et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96 (2013).
Abernathy, D. G. et al. MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 21, 332–348.e9 (2017).
Lee, S. W., Oh, Y. M., Lu, Y. L., Kim, W. K. & Yoo, A. S. MicroRNAs overcome cell fate barrier by reducing EZH2-controlled REST stability during neuronal conversion of human adult fibroblasts. Dev. Cell 46, 73–84.e7 (2018).
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C. & Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005).
Conaco, C., Otto, S., Han, J. J. & Mandel, G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl Acad. Sci. USA 103, 2422–2427 (2006).
Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18 (2018).
Adachi, K. et al. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell 23, 266–275.e6 (2018).
Ang, C. E. et al. The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. eLife 8, e41770 (2019).
Loewer, S. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113–1117 (2010).
Wang, Y. et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25, 69–80 (2013).
Bao, X. et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 25, 80–92 (2015).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011).
Ng, S.-Y., Johnson, R. & Stanton, L. W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533 (2012).
Jain, A. K. et al. LncPRESS1 Is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell 64, 967–981 (2016).
Dinger, M. E. et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445 (2008).
Zheng, X. et al. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 5, e13374 (2016).
Cliff, T. S. & Dalton, S. Metabolic switching and cell fate decisions: implications for pluripotency, reprogramming and development. Curr. Opin. Genet. Dev. 46, 44–49 (2017).
Mathieu, J. & Ruohola-Baker, H. Metabolic remodeling during the loss and acquisition of pluripotency. Development 144, 541–551 (2017).
Gascón, S. et al. Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell 18, 396–409 (2016). Gascón et al. revealed a critical metabolic checkpoint important for successful murine neuronal reprogramming in vivo and in vitro.
Magistretti, P. J. & Allaman, I. A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 (2015).
Lopaschuk, G. D. & Jaswal, J. S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 56, 130–140 (2010).
Zhou, Y. et al. Comparative gene expression analyses reveal distinct molecular signatures between differentially reprogrammed cardiomyocytes. Cell Rep. 20, 3014–3024 (2017).
Li, X. et al. Mitochondrial ROS, uncoupled from ATP synthesis, determine endothelial activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells. Can. J. Physiol. Pharmacol. 95, 247–252 (2016).
Finkel, T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 287, 4434–4440 (2012).
West, A. P. et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 (2011).
Wang, X. et al. Selenium augments microRNA directed reprogramming of fibroblasts to cardiomyocytes via nanog. Sci. Rep. 6, 23017 (2016).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Hwang, B., Lee, J. H. & Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 50, 96 (2018).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Kiselev, V. Y., Andrews, T. S. & Hemberg, M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019).
Mohamed, T. M. A. et al. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104–116.e12 (2018).
Xin, M. et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl Acad. Sci. USA 110, 13839–13844 (2013).
Leach, J. P. et al. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264 (2017).
Heallen, T. et al. Hippo pathway inhibits wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461 (2011).
Trapnell, C. Defining cell types and states with single-cell genomics. Genome Res. 25, 1491–1498 (2015).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019). Saelens et al. comprehensively evaluated the performance of 45 single-cell trajectory inference methods on 110 real and 229 synthetic datasets and provide guidelines for method selection in direct reprogramming.
Buganim, Y. et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209–1222 (2012).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The human cell atlas: from vision to reality. Nature 550, 451–453 (2017).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Stegle, O., Teichmann, S. A. & Marioni, J. C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 16, 133–145 (2015).
Grün, D. et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015).
Jia, C. et al. Accounting for technical noise in differential expression analysis of single-cell RNA sequencing data. Nucleic Acids Res. 45, 10978–10988 (2017).
Mahata, B. et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids De Novo to contribute to immune homeostasis. Cell Rep. 7, 1130–1142 (2014).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Buenrostro, J. D. et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548.e16 (2018).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216.e7 (2019).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887.e17 (2019).
Welch, J. D., Hartemink, A. J. & Prins, J. F. MATCHER: manifold alignment reveals correspondence between single cell transcriptome and epigenome dynamics. Genome Biol. 18, 138 (2017).
Cheow, L. F. et al. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 13, 833–836 (2016).
Eraslan, G., Avsec, Ž., Gagneur, J. & Theis, F. J. Deep learning: new computational modelling techniques for genomics. Nat. Rev. Genet. 20, 389–403 (2019).
Zhou, H. et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 31, 1770–1783 (2017).
Tang, Y., Liu, M. L., Zang, T. & Zhang, C. L. Direct reprogramming rather than iPSC-based reprogramming maintains aging hallmarks in human motor neurons. Front. Mol. Neurosci. 10, 359 (2017).
Fulton, D. L. et al. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 10, R29 (2009).
Rackham, O. J. L. et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 48, 331–335 (2016).
D’Alessio, A. C. et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 5, 763–775 (2015).
Forrest, A. R. R. et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Franceschini, A. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2012).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2014).
Karemaker, I. D. & Vermeulen, M. Single-cell DNA methylation profiling: technologies and biological applications. Trends Biotechnol. 36, 952–965 (2018).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Pott, S. & Lieb, J. D. Single-cell ATAC-seq: strength in numbers. Genome Biol. 16, 172 (2015).
Li, G. et al. Joint profiling of DNA methylation and chromatin architecture in single cells. Nat. Methods 16, 991–993 (2019).
Ramani, V. et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017).
Zhu, C. et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 26, 1063–1070 (2019).
Lee, D. S. et al. Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nat. Methods 16, 999–1006 (2019).
Spektor, R., Tippens, N. D., Mimoso, C. A. & Soloway, P. D. Methyl-ATAC-seq measures DNA methylation at accessible chromatin. Genome Res. 29, 969–977 (2019).
Acknowledgements
We sincerely apologize to those whose work may not have been cited owing to space constraints. J.L. is supported by NIH/NHLBI R01HL139880, R01HL139976; L.Q. is supported by AHA 18TPA34180058, NIH/NHLBI R01HL128331, R01HL144551.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Molecular Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
FANTOM5: http://fantom.gsc.riken.jp/5/
Mogrify: http://www.mogrify.net/
STRING: https://string-db.org/
Supplementary information
Glossary
- Myoblasts
-
The embryonic precursors of myocytes (muscle cells).
- Eosinophils
-
A type of white blood cell containing granules that could be intensively stained with eosin.
- Embryonic germ layer
-
A layer of cells that form at the early stages of embryonic development. The three embryonic germ layers are the endoderm, ectoderm and mesoderm. Cells in each germ layer interact with each other and differentiate to form tissues and embryonic organs.
- Autologous cell therapies
-
A novel therapeutic intervention that utilizes patients’ own cells to obtain therapeutic cells through ex vivo differentiation or reprogramming.
- Microenvironment
-
The surrounding environment of a cell that contains chemical and physical signals that directly or indirectly regulate cellular behaviour.
- Macroglial cells
-
The non-neuronal cells that provide support and protection for neurons.
- Sendai virus
-
A single strand, negative-sense RNA virus that has a large capacity for gene expression and a wide host range.
- Single guide RNA
-
RNA molecule that contains a short sequence complementary to the target DNA sequence and is used to direct Cas9 endonuclease to target loci.
- Hydrogel
-
A network of polymer chains that are hydrophilic and has been extensively studied as a scaffold for in vivo drug delivery.
- Poised enhancers
-
A subclass of enhancers enriched for both the active and the repressive histone marks. In pluripotent cells, these poised enhancers are located near key early developmental genes and are primed to activate target gene expression upon the right environmental cues.
- DNA methyltransferase Dnmt3a
-
Dnmt3a is an enzyme that catalyses the addition of methyl groups to unmethylated DNA at specific CpG sites.
- Epithelial−mesenchymal transition
-
The process where polarized epithelial cells are transformed into mobile and extracellular matrix-secreting mesenchymal cells.
- Oxidative phosphorylation
-
(OxPhos). A process in which ATP is produced because of electron transfer in mitochondria. OxPhos is the main energy source for cells like neurons, cardiomyocytes or muscle-skeletal cells.
- Reactive oxygen species
-
(ROS). A natural by-product of the electron transport chain during the oxidation of glucose. ROS can act as signalling molecules; high levels of ROS can lead to oxidative stress in a cell.
- Glycolysis
-
A metabolic pathway that converts glucose to pyruvate and produces two ATPs. In proliferating cells, glycolysis is a major resource for energy and macromolecules for biosynthesis.
- Microfluidics
-
The precise control and manipulation of fluids at a small scale.
- RNA velocity
-
The time derivative of the gene expression state. It can be used to predict the future state of individual cells on a timescale of hours.
- CpG islands
-
Short interspersed DNA sequences that are 1,000 base pairs on average and show an unusually elevated level of CpG dinucleotides. Most of the CpG islands are found at gene promoters.
Rights and permissions
About this article
Cite this article
Wang, H., Yang, Y., Liu, J. et al. Direct cell reprogramming: approaches, mechanisms and progress. Nat Rev Mol Cell Biol 22, 410–424 (2021). https://doi.org/10.1038/s41580-021-00335-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-021-00335-z
This article is cited by
-
Rapid and high-purity differentiation of human medium spiny neurons reveals LMNB1 hypofunction and subtype necessity in modeling Huntington’s disease
Inflammation and Regeneration (2024)
-
Partial reprogramming of the mammalian brain
Nature Aging (2024)
-
Decoding the basis of histological variation in human cancer
Nature Reviews Cancer (2024)
-
Cell-fate conversion of intestinal cells in adult Drosophila midgut by depleting a single transcription factor
Nature Communications (2024)
-
Cellular reprogramming as a tool to model human aging in a dish
Nature Communications (2024)