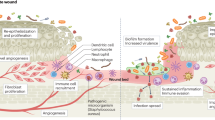
Abstract
The coagulase-negative bacterium Staphylococcus epidermidis is a member of the human skin microbiota. S. epidermidis is not merely a passive resident on skin but actively primes the cutaneous immune response, maintains skin homeostasis and prevents opportunistic pathogens from causing disease via colonization resistance. However, it is now appreciated that S. epidermidis and its interactions with the host exist on a spectrum of potential pathogenicity derived from its high strain-level heterogeneity. S. epidermidis is the most common cause of implant-associated infections and is a canonical opportunistic biofilm former. Additional emerging evidence suggests that some strains of S. epidermidis may contribute to the pathogenesis of common skin diseases. Here, we highlight new developments in our understanding of S. epidermidis strain diversity, skin colonization dynamics and its multifaceted interactions with the host and other members of the skin microbiota.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 (2018).
Gallo, R. L. Human skin is the largest epithelial surface for interaction with microbes. J. Invest. Dermatol. 137, 1213–1214 (2017). This perspective highlights the previously underappreciated large surface area of skin.
Bay, L. et al. Universal dermal microbiome in human skin. mBio 11, e02945-19 (2020).
Nakatsuji, T. et al. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 4, 1431 (2013).
Scharschmidt, T. C. et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015).
Naik, S. et al. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 143, 26–35 (2018).
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012).
Meisel, J. S. et al. Commensal microbiota modulate gene expression in the skin. Microbiome 6, 20 (2018).
Leshem, A., Liwinski, T. & Elinav, E. Immune–microbiota interplay and colonization resistance in infection. Mol. Cell 78, 597–613 (2020).
Parlet, C. P., Brown, M. M. & Horswill, A. R. Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol. 27, 497–507 (2019).
Del Rosso, J., Zeichner, J., Alexis, A., Cohen, D. & Berson, D. Understanding the epidermal barrier in healthy and compromised skin: clinically relevant information for the dermatology practitioner. J. Clin. Aesthet. Dermatol. 9, 2–8 (2011).
Kloos, W. E. & Schleifer, K. H. Isolation and characterization of staphylococci from human skin. Descriptions of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int. J. Syst. Bacteriol. 25, 62–79 (1975).
Becker, K., Heilmann, C. & Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27, 870–926 (2014).
Fleer, A. & Verhoef, J. New aspects of staphylococcal infections: emergence of coagulase-negative staphylococci as pathogens. Antonie van Leeuwenhoek 50, 72–744 (1984).
Rendboe, A. K. et al. The epidome — a species-specific approach to assess the population structure and heterogeneity of Staphylococcus epidermidis colonization and infection. BMC Microbiol. 20, 362 (2020).
Lamers, R. P. et al. Phylogenetic relationships among Staphylococcus species and refinement of cluster groups based on multilocus data. BMC Evol. Biol. 12, 171 (2012).
Brown, M. M. & Horswill, A. R. Staphylococcus epidermidis — skin friend or foe? PloS Pathog. 16, e1009026 (2020).
Otto, M. Staphylococcus epidermidis — the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567 (2009).
Milisavljevic, V. et al. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am. J. Infect. Control. 33, 341–347 (2005).
Vuong, C. & Otto, M. Staphylococcus epidermidis infections. Microbes Infect. 4, 481–489 (2002).
Oliveira, W. F. et al. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect. 98, 111–117 (2018).
Lee, J. Y. H. et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 3, 1175–1185 (2018).
Nakatsuji, T., Cheng, J. Y. & Gallo, R. L. Mechanisms for control of skin immune function by the microbiome. Curr. Opin. Immunol. 72, 324–330 (2021).
Nakatsuji, T. et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat. Med. 27, 700–709 (2021). This study demonstrates the utility of commensal staphylococci as ‘biotherapeutics’ for the common skin ailment atopic dermatitis.
Chen, Y. E., Fischbach, M. A. & Belkaid, Y. Skin microbiota–host interactions. Nature 533, 427–436 (2018).
Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
Oh, J. et al. Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016).
Kong, H. H. et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012).
Foster, T. J. Surface proteins of Staphylococcus epidermidis. Front. Microbiol. 11, 1829 (2020).
Roy, P., Horswill, A. R. & Fey, P. D. Glycan-dependent corneocyte adherence of Staphylococcus epidermidis mediated by the lectin subdomain of Aap. mBio 12, e02908–e02920 (2021). This study identifies the specific proteinaceous interaction between S. epidermidis Aap and human corneocytes necessary for colonization of the stratum corneum.
Macintosh, R. L. et al. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol. 191, 7007–7016 (2009).
Foster, T. J. The MSCRAMM family of cell-wall-anchored surface proteins of Gram-positive cocci. Trends Microbiol. 27, 927–941 (2019).
Trivedi, S. et al. The surface protein SdrF mediates Staphylococcus epidermidis adherence to keratin. J. Infect. Dis. 215, 1846–1854 (2017).
Mccrea, K. W. et al. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146, 1535–1546 (2000).
Büttner, H. et al. A giant extracellular matrix binding protein of Staphylococcus epidermidis binds surface-immobilized fibronectin via a novel mechanism. mBio 11, e01612–e01620 (2020).
Hussain, M., Heilmann, C., Peters, G. & Herrmann, M. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb. Pathog. 31, 261–270 (2001).
Leech, J. M. et al. Toxin-triggered interleukin-1 receptor signaling enables early-life discrimination of pathogenic versus commensal skin bacteria. Cell Host Microbe 26, 1–15 (2019).
Ali, N. & Rosenblum, M. D. Regulatory T cells in skin. Immunology 152, 372–381 (2017).
Scharschmidt, T. C. et al. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell Host Microbe 21, 467–477.e5 (2017).
Constantinides, M. G. et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019). This study identifies that crosstalk between non-classical immune cells and the microbiota is important for tissue development and wound healing.
Naik, S. et al. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015).
Linehan, J. L. et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796.e18 (2018).
Volz, T. et al. Induction of IL-10-balanced immune profiles following exposure to LTA from Staphylococcus epidermidis. Exp. Dermatol. 27, 318–326 (2018).
Uberoi, A. et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 29, 1235–1248 (2021). This study demonstrates that a defined microbial consortia was sufficient to regulate skin barrier integrity and repair.
Zheng, Y. et al. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe 30, 1–13 (2022). This study identifies a specific S. epidermidis molecular product that protects skin from dehydration.
Di Domizio, J. et al. The commensal skin microbiota triggers type I IFN-dependent innate repair responses in injured skin. Nat. Immunol. 21, 1034–1045 (2020).
Harrison, O. J. et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019).
Belmesk, L. et al. Prominent role of type 2 immunity in skin diseases — beyond atopic dermatitis. J. Cutan. Med. Surg. 26, 33–49 (2021).
Kim, J., Kim, B. E. & Leung, D. Y. M. Pathophysiology of atopic dermatitis: clinical implications. Allergy Asthma Proc. 40, 84–92 (2019).
Luqman, A. et al. Trace amines produced by skin bacteria accelerate wound healing in mice. Commun. Biol. 3, 277 (2020).
Nakatsuji, T. et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci. Adv. 4, eaao4502 (2018).
Keshari, S. et al. Butyric acid from probiotic Staphylococcus epidermidis in the skin microbiome down-regulates the ultraviolet-induced pro-inflammatory IL-6 cytokine via short-chain fatty acid receptor. Int. J. Mol. Sci. 20, 4477 (2019).
Li, D. et al. Lipopeptide 78 from Staphylococcus epidermidis activates β-catenin to inhibit skin inflammation. J. Immunol. 202, 1219–1228 (2019).
Lai, Y. et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 12, 1377–1382 (2009).
García-Gómez, E. et al. Staphylococcus epidermidis lipoteichoic acid: exocellular release and ltaS gene expression in clinical and commensal isolates. J. Med. Microbiol. 66, 864–873 (2017).
Hersh, A. L., Chambers, H. F., Maselli, J. H. & Gonzales, R. National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch. Intern. Med. 168, 1585–1591 (2008).
Craft, K. M., Nguyen, J. M., Berg, L. J. & Townsend, S. D. Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Medchemcomm 10, 1231–1241 (2019).
Wanke, I. et al. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Invest. Dermatol. 131, 382–390 (2011).
Bitschar, K. et al. Staphylococcus aureus skin colonization is enhanced by the interaction of neutrophil extracellular traps with keratinocytes. J. Invest. Dermatol. 140, 1054–1065.e4 (2020).
Rademacher, F. et al. Staphylococcus epidermidis activates aryl hydrocarbon receptor signaling in human keratinocytes: implications for cutaneous defense. J. Innate Immun. 11, 125–135 (2019).
Burian, M., Bitschar, K., Dylus, B., Peschel, A. & Schittek, B. The protective effect of microbiota on S. aureus skin colonization depends on the integrity of the epithelial barrier. J. Invest. Dermatol. 137, 976–979 (2017). This short report demonstrates the dual importance of S. epidermidis colonization and barrier integrity in resisting S. aureus skin colonization.
Peschel, A. & Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673 (2013).
Cogen, A. L. et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200 (2010).
Cogen, A. L. et al. Staphylococcus epidermidis antimicrobial δ-toxin (phenol-soluble modulin-γ) cooperates with host antimicrobial peptides to kill group A Streptococcus. PloS ONE 5, e8557 (2010).
Williams, M. R. et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl Med. 11, eaat8329 (2019).
Byrd, A. L. et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl Med. 9, eaal4651 (2017). This study is one of the first to identify that S. epidermidis, akin to S. aureus, can expand in some lesional atopic dermatitis sites.
Saxena, R. et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Front. Cell. Infect. Microbiol. 8, 364 (2018).
Xu, Z. et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 6, 24877 (2016).
Sanders, M. G. H., Nijsten, T., Verlouw, J., Kraaij, R. & Pardo, L. M. Composition of cutaneous bacterial microbiome in seborrheic dermatitis patients: a cross-sectional study. PloS ONE 16, e0251136 (2021).
Tanaka, A. et al. Comprehensive pyrosequencing analysis of the bacterial microbiota of the skin of patients with seborrheic dermatitis. Microbiol. Immunol. 60, 521–526 (2016).
Woo, Y. R., Lee, S. H., Cho, S. H., Lee, J. D. & Kim, H. S. Characterization and analysis of the skin microbiota in rosacea: impact of systemic antibiotics. J. Clin. Med. 9, 185 (2020).
Holmes, A. D. Potential role of microorganisms in the pathogenesis of rosacea. J. Am. Acad. Dermatol. 69, 1025–1032 (2013).
Kim, H. S. Microbiota in rosacea. Am. J. Clin. Dermatol. 21, 25–35 (2020).
Schilcher, K. & Horswill, A. R. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 84, e00026-19 (2020).
Joubert, I. A., Otto, M., Strunk, T. & Currie, A. J. Look who’s talking: host and pathogen drivers of Staphylococcus epidermidis virulence in neonatal sepsis. Int. J. Mol. Sci. 23, 860 (2022).
Both, A. et al. Distinct clonal lineages and within-host diversification shape invasive Staphylococcus epidermidis populations. PloS Pathog. 17, e1009304 (2021). This study utilizes patient-matched isolates to demonstrate S. epidermidis genetic flexibility and adaptation to various environments.
Miragaia, M., Thomas, J. C., Couto, I., Enright, M. C. & De Lencastre, H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189, 2540–2552 (2007).
Mendes, R. E., Deshpande, L. M., Costello, A. J. & Farrell, D. J. Molecular epidemiology of Staphylococcus epidermidis clinical isolates from US hospitals. Antimicrob. Agents Chemother. 56, 4656–4661 (2012).
Conlan, S. et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64 (2012). This study increases the number of publicly available S. epidermidis draft genomes and highlights genetic differences between commensal and nosocomial isolates.
Du, X. et al. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle. Nat. Microbiol. 6, 757–768 (2021).
Fišarová, L. et al. Staphylococcus epidermidis phages transduce antimicrobial resistance plasmids and mobilize chromosomal islands. mSphere 6, e00223-21 (2021).
Thakuria, B. & Lahon, K. The β-lactam antibiotics as an empirical therapy in a developing country: an update on their current status and recommendations to counter the resistance against them. J. Clin. Diagn. Res. 7, 1207–1214 (2013).
Ray, M. D., Boundy, S. & Archer, G. L. Transfer of the methicillin resistance genomic island among staphylococci by conjugation. Mol. Microbiol. 100, 675–685 (2016).
Krediet, T. G. et al. Molecular epidemiology of coagulase-negative staphylococci causing sepsis in a neonatal intensive care unit over an 11-year period. J. Clin. Microbiol. 42, 992–995 (2004).
Månsson, E. et al. Methicillin-resistant Staphylococcus epidermidis lineages in the nasal and skin microbiota of patients planned for arthroplasty surgery. Microorganisms 9, 1–14 (2021).
Mé Ric, G. et al. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol. Evol. 7, 1313–1328 (2015).
Datta, M. S. et al. Rapid methicillin resistance diversification in Staphylococcus epidermidis colonizing human neonates. Nat. Commun. 12, 6062 (2021).
Salgueiro, V. C. et al. High rate of neonates colonized by methicillin-resistant Staphylococcus species in an intensive care unit. J. Infect. Dev. Ctries 13, 810–816 (2019).
Barbier, F. et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec Iva between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202, 270–281 (2010).
Miragaia, M. et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PloS ONE 4, e7722 (2009).
Hung, W. C. et al. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PloS ONE 10, e0143106 (2015).
Banaszkiewicz, S. et al. Genetic diversity of composite enterotoxigenic Staphylococcus epidermidis pathogenicity islands. Genome Biol. Evol. 11, 3498–3509 (2019).
Argemi, X., Hansmann, Y., Prola, K. & Prévost, G. Coagulase-negative staphylococci pathogenomics. Int. J. Mol. Sci. 20, 1215 (2019).
Zhou, W. et al. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell 180, 454–470.e18 (2020). This study is the first longitudinal, multi-patient assessment of S. epidermidis strain-level diversity on healthy skin.
Méric, G. et al. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat. Commun. 9, 5034 (2018).
Harris, S. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 466–469 (2010).
Williams, M. R. et al. Interplay of staphylococcal and host proteases promotes skin barrier disruption in Netherton syndrome. Cell Rep. 30, 2923–2933.e7 (2020).
Cau, L. et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J. Allergy Clin. Immunol. 147, 955–966.e16 (2021). This study implicates the production of the S. epidermidis EcpA protease in the disruption and exacerbation of atopic skin lesions.
Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228 (2008).
Paharik, A. E. & Horswill, A. R. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol. Spectr. 4, 529–566 (2016).
Schaeffer, C. R. et al. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 83, 214–226 (2015).
Schaeffer, C. R. et al. Versatility of biofilm matrix molecules in Staphylococcus epidermidis clinical isolates and importance of polysaccharide intercellular adhesin expression during high shear stress. mSphere 1, e00165-16 (2016).
Fey, P. D. & Olson, M. E. Current concepts in biofilm formation of Staphylococcus epidermidis. Fut. Microbiol. 5, 917–933 (2010).
O’Gara, J. P. Into the storm: chasing the opportunistic pathogen Staphylococcus aureus from skin colonisation to life-threatening infections. Environ. Microbiol. 19, 3823–3833 (2017).
Kozitskaya, S. et al. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43, 4751–4757 (2005).
Kozitskaya, S. et al. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72, 1210–1215 (2004).
Rogers, K. L., Rupp, M. E. & Fey, P. D. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl. Environ. Microbiol. 74, 6155–6157 (2008).
Paharik, A. E. et al. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol. Microbiol. 103, 860–874 (2017).
Rohde, H. et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895 (2005).
Conrady, D. G. et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl Acad. Sci. USA 105, 19456–19461 (2008).
Formosa-Dague, C., Speziale, P., Foster, T. J., Geoghegan, J. A. & Dufrêne, Y. F. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl Acad. Sci. USA 113, 410–415 (2016).
Salava, A. & Lauerma, A. Role of the skin microbiome in atopic dermatitis. Clin. Transl Allergy 4, 1–6 (2014).
Meylan, P. et al. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J. Invest. Dermatol. 137, 2497–2504 (2017).
Allen, H. B. et al. The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol. 150, 260–265 (2014).
Allen, H. B., Mueller, J. L. & Herbert Allen, C. B. A novel finding in atopic dermatitis: film-producing Staphylococcus epidermidis as an etiology. Int. J. Dermatol. 50, 992–993 (2011).
Gonzalez, T. et al. Biofilm propensity of Staphylococcus aureus skin isolates is associated with increased atopic dermatitis severity and barrier dysfunction in the MPAACH pediatric cohort. Allergy 76, 302–313 (2021). This work presents a clinically robust and mechanistic assessment of paediatric atopic dermatitis development and progression in a US cohort.
Yarwood, J. M., Bartels, D. J., Volper, E. M. & Greenberg, E. P. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850 (2004).
Kavanaugh, J. S. & Horswill, A. R. Impact of environmental cues on staphylococcal quorum sensing and biofilm development. J. Biol. Chem. 291, 12556–12564 (2016).
Thoendel, M., Kavanaugh, J. S., Flack, C. E. & Horswill, A. R. Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151 (2011).
Novick, R. P. & Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008).
Yang, T., Tal-Gan, Y., Paharik, A. E., Horswill, A. R. & Blackwell, H. E. Structure–function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem. Biol. 11, 1982–1991 (2016).
Wright, J. S., Jin, R. & Novick, R. P. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl Acad. Sci. USA 102, 1691–1696 (2005).
Nakamura, Y. et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci. Transl Med. 12, eaay4068 (2020).
Otto, M., Echner, H., Voelter, W. & Götz, F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69, 1957–1960 (2001).
Brown, M. M. et al. Novel peptide from commensal Staphylococcus simulans blocks methicillin-resistant Staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob. Agents Chemother. 64, e00172-20 (2020).
Khan, B. A., Yeh, A. J., Cheung, G. Y. & Otto, M. Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 24, 689–704 (2015).
Harraghy, N., Kerdudou, S. & Herrmann, M. Quorum-sensing systems in staphylococci as therapeutic targets. Anal. Bioanal. Chem. 387, 437–444 (2007).
Olson, M. E. et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196, 3482–3493 (2014).
Nakatsuji, T. et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl Med. 9, eaah4680 (2017).
Ebner, P. et al. Lantibiotic production is a burden for the producing staphylococci. Sci. Rep. 8, 7471 (2018).
Heilbronner, S., Krismer, B., Brötz-Oesterhelt, H. & Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 19, 726–739 (2021).
Liu, Y. et al. Skin microbiota analysis-inspired development of novel anti-infectives. Microbiome 8, 85 (2020).
O’Sullivan, J. N., Rea, M. C., O’Connor, P. M., Hill, C. & Ross, R. P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 95, fiy241 (2019).
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
Sandiford, S. & Upton, M. Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against staphylococci. Antimicrob. Agents Chemother. 56, 1539–1547 (2012).
Janek, D., Zipperer, A., Kulik, A., Krismer, B. & Peschel, A. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PloS Pathog. 12, e1005812 (2016).
Ekkelenkamp, M. B. et al. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 579, 1917–1922 (2005).
Chin, D. et al. Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat. Commun. 12, 1887 (2021).
Claudel, J. P. et al. Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatology 235, 287–294 (2019).
Woo, T. E. & Sibley, C. D. The emerging utility of the cutaneous microbiome in the treatment of acne and atopic dermatitis. J. Am. Acad. Dermatol. 82, 222–228 (2020).
Fitz-Gibbon, S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 (2013).
Christensen, G. J. M. et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17, 152 (2016).
Nakamura, K. et al. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 10, 21237 (2020).
Claesen, J. et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl Med. 12, 5445 (2020). This study identifies a novel antimicrobial made by C. acnes in the hair follicle to facilitate inter-bacterial competition.
Wang, Y. et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 98, 411–424 (2014).
Wang, Y. et al. A precision microbiome approach using sucrose for selective augmentation of Staphylococcus epidermidis fermentation against Propionibacterium acnes. Int. J. Mol. Sci. 17, 1870 (2016).
Xia, X. et al. Staphylococcal LTA-induced miR-143 inhibits Propionibacterium acnes-mediated inflammatory response in skin. J. Invest. Dermatol. 136, 621–630 (2016).
Sakr, A., Brégeon, F., Mège, J. L., Rolain, J. M. & Blin, O. Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 9, 2419 (2018).
Totté, J. E. E. et al. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br. J. Dermatol. 175, 687–695 (2016).
Krismer, B., Weidenmaier, C., Zipperer, A. & Peschel, A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 15, 675–687 (2017).
Oh, J., Conlan, S., Polley, E. C., Segre, J. A. & Kong, H. H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 1–11 (2012).
Liu, Q. et al. Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 27, 1–11 (2019).
Iwase, T. et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010).
Sugimoto, S. et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host–pathogen interaction. J. Bacteriol. 195, 1645–1655 (2013).
Chen, C. et al. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 288, 29440–29452 (2013).
Roche, F. M., Meehan, M. & Foster, T. J. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology (Reading) 149, 2759–2767 (2003).
McLoughlin, I. J., Wright, E. M., Tagg, J. R., Jain, R. & Hale, J. D. F. Skin microbiome — the next frontier for probiotic intervention. Probiotics Antimicrob. Proteins https://doi.org/10.1007/s12602-021-09824-1 (2021).
Nodake, Y. et al. Pilot study on novel skin care method by augmentation with Staphylococcus epidermidis, an autologous skin microbe — a blinded randomized clinical trial. J. Dermatol. Sci. 79, 119–126 (2015).
Sakr, A., Brégeon, F., Rolain, J. M. & Blin, O. Staphylococcus aureus nasal decolonization strategies: a review. Expert Rev. Anti Infect. Ther. 17, 327–340 (2019).
Dodds, D. et al. Controlling the growth of the skin commensal Staphylococcus epidermidis using d-alanine auxotrophy. mSphere 5, e00360-20 (2020).
Ito, Y. et al. Staphylococcus cohnii is a potentially biotherapeutic skin commensal alleviating skin inflammation. Cell Rep. 35, 109052 (2021).
Holz, C. et al. Novel bioactive from Lactobacillus brevis DSM17250 to stimulate the growth of Staphylococcus epidermidis: a pilot study. Benef. Microbes 8, 121–131 (2017).
Williams, M. R. & Gallo, R. L. Evidence that human skin microbiome dysbiosis promotes atopic dermatitis. J. Invest. Dermatol. 137, 2460–2461 (2017).
Myles, I. A. et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 3, e120608 (2018).
Volz, T. et al. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J. Invest. Dermatol. 134, 96–104 (2014).
Kao, M. S. et al. Microbiome precision editing: using PEG as a selective fermentation initiator against methicillin-resistant Staphylococcus aureus. Biotechnol. J. 12, 1600399 (2017).
Liu-Walsh, F. et al. Prebiotic colloidal oat supports the growth of cutaneous commensal bacteria including S. epidermidis and enhances the production of lactic acid. Clin. Cosmet. Investig. Dermatol. 14, 73–82 (2021).
Götz, F., Perconti, S., Popella, P., Werner, R. & Schlag, M. Epidermin and gallidermin: staphylococcal lantibiotics. Int. J. Med. Microbiol. 304, 63–71 (2014).
Schoenfelder, S. M. K. et al. Success through diversity — how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 300, 380–386 (2010).
Ferretti, P. et al. Experimental metagenomics and ribosomal profiling of the human skin microbiome. Exp. Dermatol. 26, 211–219 (2017).
Van Rossum, T., Ferretti, P., Maistrenko, O. M. & Bork, P. Diversity within species: interpreting strains in microbiomes. Nat. Rev. Microbiol. 18, 491–506 (2020).
Dengler Haunreiter, V. et al. In-host evolution of Staphylococcus epidermidis in a pacemaker-associated endocarditis resulting in increased antibiotic tolerance. Nat. Commun. 10, 1149 (2019).
Gu, J. et al. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61, 342–348 (2005).
Lee, J. Y. H. et al. Mining the methylome reveals extensive diversity in Staphylococcus epidermidis restriction modification. mBio 10, e02451-19 (2019).
Marraffini, L. & Sontheimer, E. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Rossi, C. C., Souza-Silva, T., Araújo-Alves, A. V. & Giambiagi-deMarval, M. CRISPR–Cas systems features and the gene-reservoir role of coagulase-negative staphylococci. Front. Microbiol. 8, 1545 (2017).
Lyon, G. J. & Novick, R. P. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25, 1389–1403 (2004).
Acknowledgements
M.M.S. was supported by the National Institute of Allergy and Infectious Diseases (NIAID) Ruth L. Kirschstein National Research Award Predoctoral Fellowship AI157052. A.R.H. was supported by the NIAID grants AI153185 and AI162964 and the US Department of Veteran Affairs grant BX002711.
Author information
Authors and Affiliations
Contributions
M.M.S. researched data for the article. A.R.H. and M.M.S. contributed substantially to discussion of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Michael Otto, Wilma Ziebuhr and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Stratum corneum
-
The topmost layer of the epidermis, composed of dead keratinocytes linked together in a ‘brick and mortar’ structure by extracellular lipids.
- Coagulase
-
A polypeptide secreted by Staphylococcus aureus that promotes blood clotting and was historically used to distinguish this organism from less pathogenic coagulase-negative staphylococci (CoNS) for clinical identification.
- Sortase
-
An enzyme that links surface proteins to the Gram-positive cell wall.
- Corneocytes
-
Dead keratinocytes that compose the topmost layer of the epidermis.
- Extracellular matrix
-
A structural support system within the dermal skin layer composed of various proteins including collagen and elastin.
- Sphingomyelinase
-
A bacterial esterase, often implicated in virulence, that cleaves sphingomyelin.
- Pangenome
-
The entire set of genes within a species, including conserved, core genes found in every strain and variable genes not conserved in every strain.
- KEGG modules
-
Families of genes that are linked to specific cellular functions.
- Phase variation
-
The reversible, heterogeneous switch in gene expression profiles within a clonal population of bacterial cells.
- Plasmacytoid dendritic cells
-
(pDCs). A specialized subpopulation of dendritic cells involved in immunosurveillance and antigen recognition.
- Corneodesmosomes
-
The major intercellular adhesives within the stratum corneum.
- Netherton syndrome
-
A rare genetic disorder characterized by a mutation in a serine protease inhibitor that leads to severe, often life-threatening, skin abnormalities.
- Bacteriocins
-
Ribosomally synthesized peptides made by many types of bacteria with varying mechanisms of antimicrobial activity.
- Kin selection
-
The theory that genetic relatedness of a population of cells should lead to more cooperativity and increased fitness of that specific population.
- Probiotics
-
Live microorganisms applied to re-regulate a dysbiotic community or actively exclude an opportunistic pathogen.
- Prebiotics
-
Nutrients or other substrates added to alter species composition that favour select microbial growth and exclude opportunistic pathogens.
- Postbiotic
-
Dead or otherwise inanimate bacteria or a bacterial product applied to promote skin barrier function.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Severn, M.M., Horswill, A.R. Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat Rev Microbiol 21, 97–111 (2023). https://doi.org/10.1038/s41579-022-00780-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00780-3
This article is cited by
-
Using novel micropore technology combined with artificial intelligence to differentiate Staphylococcus aureus and Staphylococcus epidermidis
Scientific Reports (2024)
-
Skin Deep: The Potential of Microbiome Cosmetics
Journal of Microbiology (2024)
-
Microbiological study of sternal osteomyelitis after median thoracotomy – a retrospective cohort study
BMC Infectious Diseases (2023)
-
The dual activity of CaONPs as a cancer treatment substance and at the same time resistance to harmful microbes
Scientific Reports (2023)
-
The bactericidal and antibiofilm effects of a lysine-substituted hybrid peptide, CM-10K14K, on biofilm-forming Staphylococcus epidermidis
Scientific Reports (2023)