Abstract
Drug repurposing is the use of a given therapeutic agent for indications other than that for which it was originally designed or intended. The concept is appealing because of potentially lower development costs and shorter timelines than are needed to produce a new drug. To date, drug repurposing for cardiovascular indications has been opportunistic and driven by knowledge of disease mechanisms or serendipitous observation rather than by systematic endeavours to match an existing drug to a new indication. Innovations in two areas of personalized medicine — computational approaches to associate drug effects with disease signatures and predictive model systems to screen drugs for disease-modifying activities — support efforts that together create an efficient pipeline to systematically repurpose drugs to treat cardiovascular disease. Furthermore, new experimental strategies that guide the medicinal chemistry re-engineering of drugs could improve repurposing efforts by tailoring a medicine to its new indication. In this Review, we summarize the historical approach to repurposing and discuss the technological advances that have created a new landscape of opportunities.
Key points
-
Contemporary technologies are expected to make drug repurposing large-scale, systematic and deliberate rather than opportunistic.
-
New experimental and computational tools harness patient genomics for drug repurposing.
-
Discovery of repurposed drugs on the basis of patient genomics has implications for precision prescribing of medicines to treat individual patients.
-
The treatment of rare, monogenic diseases, which often provide too little return on investment to incentivize conventional drug discovery, might benefit because the molecular aetiologies of these diseases are well suited to the discovery of drug repurposing candidates.
Similar content being viewed by others
Introduction
Cardiovascular disease, which includes diseases that affect the heart, vasculature and blood, is the major cause of death1. Despite this alarming statistic, the pipeline for new cardiovascular medicines has been diminishing compared with drug development in oncology, neurology and other major fields of medicine. For instance, in the 3 years from 2018 to 2020, the FDA’s Center for Drug Evaluation and Research approved record numbers of new medical entities (59, 48 and 53 in each year, respectively)2. However, only four new medical entities were approved during this period for cardiovascular indications, compared with 49 for cancer2. A primary driver of this disparity is the high cost of cardiovascular clinical trials3,4.
The diminished pipeline for new cardiovascular medicines has spurred interest in technologies that can be used to repurpose drugs to serve the unmet needs of patients. Drug repurposing (also called drug repositioning, re-profiling or re‐tasking) is the strategy used to discover new applications for approved, abandoned or investigational drugs that are outside the scope of the originally intended indication of the drug. The strategy has several advantages over developing entirely new drugs5. Notably, the time frame for development and the risk of failure are lower because the existing compound will have been shown to be safe in preclinical models and in humans (if clinical testing is sufficiently advanced). Although drug repurposing will not reverse the decline in the cardiovascular drug pipeline, this approach could potentially decrease the cost of a new drug, depending on the stage of clinical development and regulatory approval.
The fundamental premise of repurposing is that drugs have pleiotropic effects beyond their known mechanism of action. Although this phenomenon is apparent from the clinical off-target effects of drugs, it has also been confirmed at the transcriptional level by the NIH Library of Integrated Network-Based Cellular Signatures (LINCS) Program by profiling changes in gene expression induced by drug treatment. A drug is matched to a disease not only by its known drug–target interaction but also by its diverse effects on multiple genes, mRNAs and proteins. A fascinating manifestation of this principle is that some medications are of clinical benefit contrary to expectations based on their known mechanism of action. For example, angiotensin-converting enzyme inhibitors increase survival in patients with hypertension even though angiotensin levels return to their pre-medication levels after 18 months of ongoing treatment6.
Although drug repurposing as a concept is not new, this approach has historically been largely opportunistic rather than deliberate5, but owing to technological advances, the effects of drugs can now be prospectively matched to new indications. The new strategies rely on extensive phenotyping of a drug’s effect at the level of patient care gleaned from electronic health records and, increasingly, on the identification of drugs on the basis of their effects on proteins, cell signalling and gene expression and their inverse relationship with molecular phenotypes of disease (Box 1). Complementing these approaches, high-throughput screening using realistic model systems can be used to identify compounds that ameliorate disease phenotypes, without prior knowledge of the intended targets of the drugs. Given that both strategies match drugs to the underlying patient genetics and/or molecular mechanisms of disease, they comprise a systematic and mechanism-based approach to drug repurposing.
The emphasis on therapeutic mechanisms increases the precision of drug discovery, just as mechanism-based diagnosis and treatment has influenced clinical practice. Moreover, matching compounds to molecular mechanisms is likely to expand the number of indications that can be treated and increase the precision of clinical care. The mechanistic focus should help to address rare cardiovascular indications, for which the return on investment has often been insufficient motivation to develop a drug from concept to bedside. In aggregate, rare cardiovascular diseases are estimated to affect up to 3% of the population7,8, making the lack of drug development for this population a major health problem. Rare diseases often have monogenic aetiologies7,8, for which the clear genetics make them well suited to new computational and experimental drug discovery technologies.
In this Review, we provide background on conventional approaches to drug repurposing and summarize newly developed techniques for systematic drug repurposing. Given that the new technologies can be used not only to repurpose drugs, but also to guide their medicinal chemistry refinement to suit their new roles, drug re-engineering is also discussed. Our view is that the adoption of these new tools will greatly accelerate the pace of drug repurposing and increase the efficiency of drug discovery for the treatment of cardiovascular disease.
Observation-driven drug repurposing
Historically, the most common approach to drug repurposing has been on the basis of astute observations made by practitioners during clinical trials or routine clinical practice. Table 1 lists examples of drug repurposing for cardiovascular and other indications9. A well-known example is the use of minoxidil to promote hair growth5. Although increased hair growth might initially have been considered an unwanted effect of treatment with minoxidil for arterial hypertension, the drug had an estimated annual worldwide sale of US $860 million in 2020 (predicted to reach more than $1 billion by the end of 2024) as a treatment for baldness10.
Perhaps the earliest examples of drug repurposing relate to applications of aspirin11. Salicylate-containing willow bark was known to have medicinal qualities long before 1000 B.C.E. The active ingredient was purified and then manufactured synthetically in the nineteenth century, primarily to be used as an antipyretic and analgesic. The effect of acetylsalicylic acid on platelet aggregation was observed in 1950, and its role in the inhibition of prostaglandin synthesis was defined in 1969. In the ensuing years, these observations led to the repurposing of aspirin to be used to inhibit platelet aggregation. Aspirin was also used to slow the progression of thrombosis in myocardial infarction and in patients with Kawasaki disease accompanied by coronary artery aneurysms11.
Over the past 20 years, elucidation of the role of inflammation — and mast cell degranulation in particular — in heart disease has prompted the exploration of other anti-inflammatory medications. For example, retrospective analyses and clinical trials suggest that histamine receptor blockade is beneficial in the context of heart failure12,13,14. For similar reasons, the anti-inflammatory agent colchicine has been explored as a treatment for coronary artery disease and myocardial infarction, including two sizeable clinical trials with mixed results15,16,17.
Assessing other important features of heart pathophysiology has led to drug repurposing efforts beyond inflammation. For example, hydralazine has antioxidant effects and reduces apoptosis, in addition to antihypertensive and afterload-reducing effects. Understanding that these processes have important roles in reperfusion injury after myocardial infarction led to the evaluation of hydralazine for the treatment of myocardial injury after infarction, with promising results in in vivo models18. Other cardiovascular drugs that have been studied as therapies for other diseases include diuretics as antiparasitic agents (tested in vitro)19, statins as adjuvant therapy for cancers (reviewed previously20) and antihypertensive agents for bipolar disorder21.
The preceding strategies focused largely on exploiting similarities between a drug’s effect and a desired outcome in a specific disease. Exploiting the clinically observed effect on patients is a major strength but contrasts with the challenge of systematization (Table 2). Another approach is to find diseases with similar pathophysiological or pathogenic features and then to explore crossover of therapies from one disease to the other. This approach was taken to identify nintedanib, an FDA-approved treatment for idiopathic pulmonary fibrosis, as a potential therapy to reduce pathological fibrosis and remodelling of the heart after myocardial infarction22. Although preclinical in vivo studies were promising22, clinical efficacy has yet to be validated.
The discovery of synergistic drug combinations can lead to novel therapeutic strategies using existing approved drugs. Analyses can take the form of in vitro screening of drug targets cross-referenced to known signalling pathways contributing to disease. This approach was used to identify the combination of allethrin and menthol as a potential therapy for heart failure, a finding that has been validated in vitro23.
Health-care centres globally are rapidly accumulating massive amounts of clinical data in their electronic health record systems. To the extent that security and privacy concerns permit, these repositories enable the validation of drug repurposing candidates by means of in silico clinical trials. Although this term is sometimes used to refer to the computational simulation of patients and/or disease processes, we are referring to the computational analysis of outcomes for one disease in response to drugs being used to treat another. Suitable cohorts of ‘treated’ and ‘control’ patients with a disease of interest can be identified in the electronic health record system, making it possible to determine whether the novel drug–disease pairing has beneficial effects on that disease. These in silico trials have obvious limitations, such as their retrospective nature, the possibly confounding influence of multiple diseases in an individual patient, and technical challenges inherent to data mining (for example, non-standardized terminology and difficulties in querying free-text data elements). Machine-learning techniques of natural language processing algorithms are an important technology being applied to overcome these difficulties24. Despite these challenges, the approach is a useful step towards full clinical validation of novel indications for existing drugs.
In addition, formal clinical trials can yield serendipitous results. Given that diabetes mellitus is a well-established risk factor for cardiovascular disease, it was not surprising when large clinical trials of diabetes-controlling medications showed a favourable effect on cardiovascular disease. What was surprising, however, was that some of these trials showed an additive beneficial effect on cardiovascular disease that was independent of the degree of glucose control25. This finding has led to extensive efforts to explore the utility of drugs such as metformin, glucagon-like peptide 1 receptor (GLP1R) agonists and sodium–glucose cotransporter 2 (SGLT2) inhibitors as treatments for hypertrophic cardiomyopathy, heart failure and coronary artery disease. Although consensus has yet to emerge about metformin and GLP1R agonists, results from numerous smaller clinical trials are promising25, indicating the need for large-scale clinical trials. Two SGLT2 inhibitors, dapagliflozin and empagliflozin, have been approved in Europe and the USA for the treatment of heart failure with reduced ejection fraction (Table 1). Interestingly, the increase in global revenue for SGLT2 inhibitors is predicted to grow immensely, spurred in part by the approved indication for heart failure, from US $7.2 billion in 2020 to $12 billion in 2026 (ref.26).
Drugs under evaluation in phase III clinical trials represent major investments, and the clinical effects have to be thoroughly characterized. Therefore, when a primary end point is met, prespecified secondary end points are analysed, often for outcomes beyond those related to the primary end point. Moreover, the delineation of effect modifiers, which can be apparent from differential effects in particular subgroups, can reveal effects that are hidden in the whole cohort. The effect of statins on cancer20 and the effect of SGLT2 inhibitors on heart failure27,28 are examples of drug repurposing based on secondary analyses of phase III clinical trials. Both have taught us the importance of well-designed secondary analyses in clinical trials, in particular making them broad enough to generate hypotheses beyond those that might be anticipated. In addition, using clinical trial databases as a resource, even ad hoc analyses of the results can generate hypotheses about alternative uses for the tested drugs. The resulting drug candidates can then be investigated for repurposing.
Clinical data, even when registered in modern electronic health records or electronic case report forms, tend to contain fields of free-text information, which are a challenge for comprehensive analysis, even with emerging technologies in artificial intelligence. Moreover, the data tend to be incomplete and to be prone to information bias. For example, information that is not required in defined text fields is less likely to be reported, even if there are free-text fields in which to report it. The lack of standardized and complete information also makes the data less useful for an agnostic approach to find a completely new target for a drug. In addition, the lack of completeness of the data makes retrospective analyses prone to confounding and false predictions (positive as well as negative).
Other biases exist. Electronic case report forms and data from randomized clinical trials come with inherent problems of generalizability because of disparities between the study population and patients treated in real life and patient selection bias. The latter is of particular concern in studies with run-in phases, which exclude poorly adherent or intolerant patients and can therefore bias outcomes in favour of a positive drug effect compared with what might be achieved in routine care. Furthermore, differences between real-life care and the care provided while patients are part of the trial can also bias findings (positively as well as negatively), with potential implications for drug repurposing.
The increasing power of information technology, including natural language processing, allows searches and analyses not only of randomized clinical trial databases but also of published literature in a systematic way and can thereby identify new correlations that can generate new hypotheses. This literature-based discovery29 relies on the assumption that two facts A and C might have a relationship to each other if they share a link, identified in the literature, with a third concept B. The term ‘ABC model’ has been coined to describe this mode of scientific discovery through inference. A practical example of this approach is the suggested use of thalidomide to treat chronic hepatitis C30. In this example, the literature search algorithm revealed that thalidomide (A) is associated with an anti-inflammatory shift in the balance of T helper (TH) cells from TH2 cells towards TH1 cells (B) and, secondly, that a bias in the opposite direction (TH1 towards TH2) occurs in chronic hepatitis C (C). The predicted therapeutic efficacy of A to C was later found to be consistent with the results from a small clinical trial31. A substantial drawback of this approach is false discoveries resulting from publication bias (the under-reporting of negative findings in the scientific literature).
Systematic drug repurposing
The ability to systematically identify novel applications for existing drugs has been greatly increased by the establishment of large-scale genomic, proteomic, transcriptomic and functional screening technologies. The fundamental tenet of these efforts is that the established mechanism of action of a given drug is potentially one of many biological effects induced by that drug. Other effects can manifest as side effects or go unnoticed in the absence of systematic assessment of the complete repertoire of effects of a given drug. Therefore, modern drug repurposing takes advantage of technologies to comprehensively match these effects to potentially therapeutic mechanisms. The methodological aspects of these computational approaches have been reviewed previously5.
Association by side effect profiles
One of the more common drug repurposing strategies has been the association of beneficial side effects of one drug with a desired outcome in another context. An example in the cardiovascular field is the use of aspirin in the context of coronary artery disease and myocardial infarction to reduce platelet aggregation, for which aspirin was not initially indicated. Furthermore, aspirin has antifibrotic effects after myocardial infarction32. However, the clinical applicability of this finding is limited by adverse effects such as bleeding and gastric ulcers that accrue with prolonged use at the dosages required for the beneficial effect. Nonetheless, the observation led to the exploration of mesalazine, which has a similar structure to that of aspirin and a better adverse effect profile, with promising in vitro results33.
The identification of beneficial side effects can be pursued systematically. One strategy has been to use the Unified Medical Language System (UMLS) ontology to match beneficial side effects to novel purposes. Applied to 746 approved drugs, researchers found 1,018 associations with strong side effects, and >25% of these could be caused by two or more chemically dissimilar compounds34. Large regulatory databases, such as the FDA Adverse Event Reporting System (FAERS), the EMA EudraVigilance and the WHO VigiBase, would be tremendous resources for further exploration in this field.
Electronic health records provide an opportunity to access large cohorts for drug effect searches, but accessibility is limited by information bias and incompleteness. For instance, the focus of a physician on one disease might lead to under-reporting of findings that seemed to be unrelated at the time of data entry. Despite these constraints, electronic health records were used to identify a reduction in cancer-related mortality in patients with diabetes who received treatment with metformin compared with those who did not receive this drug35. Observational studies have suggested that patients with diabetes and using metformin have a decreased risk of incident cancer and cancer-related mortality compared with patients not using metformin (reviewed previously36,37), and several clinical interventional studies have indicated the beneficial effect of metformin in the treatment of colorectal and ovarian cancers38,39. Importantly, however, these findings have not been confirmed in prospective, randomized trials. Although the experience with metformin has demonstrated the feasibility of this approach, it has not yet (to our knowledge) been applied to drug repurposing in the cardiovascular field.
Genome-wide association studies
The complete sequencing of the human genome enabled strategies to connect genomic sequences to phenotypic traits and diseases, with opportunities for drug repurposing40. Genome-wide association studies (GWAS) link single-nucleotide polymorphisms throughout the genome to traits by assessing linkage disequilibrium. Unlike Mendelian genetic studies, which identify rare variants with large effect sizes, GWAS take advantage of large cohort populations to link common genetic variants with relatively small effect sizes to traits. However, small effect sizes do not necessarily translate into low efficacy of a drug targeting the corresponding protein. For example, statins are highly effective, whereas the effect sizes of single-nucleotide polymorphisms in HMGCR (encoding the protein inhibited by statins) are fairly small41,42. Using GWAS, researchers showed that 92 genes encoding proteins targeted by drugs were in linkage disequilibrium with diseases other than those for which they had regulatory approval, suggesting that genes identified via GWAS can, in principle, be matched to novel targets for drugs43. Although this technology is a potential opportunity to repurpose drugs, and several have been suggested for coronary artery disease44,45, we are unaware of a clinical trial for a cardiovascular indication based on this strategy. A related approach involves mapping quantitative trait loci for circulating proteins to disease and subsequently identifying regulatory networks involving these proteins, followed by the identification of drugs that target those proteins and pathways46. To date, this type of analysis has identified multiple pathways related to cardiovascular disease that are targets of existing drugs47, but the relationships have not yet been validated.
Association by chemical structure
Candidate drugs for repurposing can also be identified by the similarity of their chemical structure to that of existing therapeutics. This approach can take the form of computational analysis of large compendia of drug structures to find the most similar candidates or simply in vitro screening of relatively small numbers of drugs with similar structures. Databases of 3D structures were established as early as the 1970s, the largest being the Cambridge Structural Database (CSD) that comprises about 500,000 structures48.
Structural matching is used to repurpose drugs by two primary means. The first is to query chemical compounds for binding to a target structure and, thereafter, to screen for effect, as has been done to identify new antituberculosis agents49. A high-throughput version of this approach was named the Similarity Ensemble Approach (SEA). Investigators predicted 184 new drug–target pairs, some with an even higher Ki than any drugs that were approved for that target50. The success rate of the predicted compounds that were tested in vitro was >70%50. A variant of this approach, the weighted ensemble similarity (WES) approach, is an algorithm to handle predictions in a complex dataset, and was used to identify the β-blocker esmolol as an agonist of peroxisome proliferator-activated receptor-γ, a finding that could potentially guide the development of bifunctional drugs to reduce cardiovascular risk beyond that achieved by blood pressure and heart rate control alone51.
The opportunities of structural biology have expanded tremendously with the increase in the spatial resolution of cryogenic electron microscopy (cryo-EM). Cryo-EM can be used to visualize proteins bound to small molecules, lipids and antibodies, thereby aiding drug discovery programmes. Different conformational states can be identified within a single dataset. The technology comes with much lower requirements for purified proteins (microgram rather than milligram quantities) than for X-ray crystallography. Although the resolution of cryo-EM now allows the visualization of detailed interactions, it still lags behind that of X-ray crystallography, and the technology is not conducive to high-throughput screening. Although cryo-EM has been used for drug repurposing, such as in the search for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) blockers52, it has not led to the repurposing of drugs in routine clinical use.
Association by transcriptomic profiling
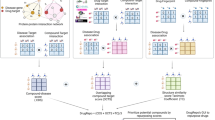
Analysis of transcriptome datasets has emerged as an important method to link drug and disease signatures53. Databases such as the NIH LINCS transcriptional library capture the effects of existing drugs and/or perturbation of specific genes on gene expression across the genome and across various cell types. By matching these transcriptomic profiles with those of diseases — or specific disease-causing genetic variants — new treatments can be identified that reverse aspects of these disease signatures (Fig. 1). When validated in vivo, advancement of FDA-approved compounds to clinical testing can be fairly quick. One of the earliest examples identified was the antiseizure medication topiramate as a potential therapeutic agent for inflammatory bowel disease54. Although a subsequent report cast doubt on the clinical utility of topiramate for inflammatory bowel disease55, the initial study is of interest because it exemplified the concept of an in silico clinical trial (discussed above).
Publicly available datasets, such as the NIH LINCS transcriptomic library, facilitate the matching of gene variant effects with drug effects. In the example depicted, defining the transcriptomes of cells treated with short hairpin RNAs to knockdown the activity of a gene of interest generates a putative signature. Cross-referencing against the transcriptional signatures of drug treatments catalogued in the database allows the identification of drugs that might reverse the gene-knockdown signature. When restricted to FDA-approved drugs, this in silico screening leads to drug repurposing candidates that can be validated via in vitro and/or in vivo models. Depending on the current indications and use of the drug in humans, the pathway to a clinical trial and clinical use might be streamlined compared with that for a completely new drug. Another avenue is to perform additional medicinal chemistry work to further optimize the activity of the drug and/or to develop novel compounds. KS, Kolmogorov–Smirnov.
To date, few examples exist of leveraging large-scale transcriptomic databases to repurpose drugs for cardiovascular applications. One example is our unpublished work relating to application of the angiotensin receptor blocker olmesartan to treat heart failure secondary to a variant in LMNA. Leveraging the LINCS database, we have established the algorithms and software tools required to identify novel therapies targeting specific diseases related to genetic variants56. We applied this technology to familial dilated cardiomyopathy caused by haploinsufficiency of LMNA (encoding lamin A/C), and our preliminary findings indicate reversal of the pathological phenotype at the single-cell level57. Whether drug repurposing by transcriptome association can be applied to other cardiomyopathies, such as hypertrophic cardiomyopathy, remains to be understood.
Off-target profiling is a logical approach for drug repurposing. Whether elucidated by transcriptional responses (as outlined above), in silico predication or direct biochemical profiling, off-target profiling is routinely performed to reveal possible side effects of compounds during the drug development process58. By the time a drug candidate has been nominated for clinical testing, knowledge of its preclinical pharmacology and its ADME (absorption, distribution, metabolism and excretion) and safety profiles can inform repurposing and provide an opportunity for clinical evaluation in unanticipated disease indications. Repurposing during the drug development process would maximize the potential value generated, not least through extension of the patent.
Association by metabolite likeness
Old drugs, such as aspirin and morphine, were originally discovered by their effect rather than their affinity for a particular target. Aspirin inhibits cyclooxygenase, and the compound (acetylsalicylic acid) can be endogenously generated in humans through the metabolism of benzoic acid. As this example suggests, known drugs can be identified as repurposing candidates on the basis of their similarity to human metabolites. By using FDA-approved, small-molecule drugs and comparing their structural similarity with a list of human metabolites, researchers applied drug–target interaction tools, such as SwissTargetPrediction59, to create an atlas of similarities between approved drugs and human metabolites60. Miglustat is an inhibitor of glucosylceramide synthetase (also known as ceramide glucosyltransferase) and is used to reduce the synthesis of glucosylceramide, which accumulates in patients with Gaucher disease, who have a defect in the enzyme β-glucosidase (also known as lysosomal acid glucosylceramidase), which converts glucosylceramide into ceramide and glucose. Based on its structural similarity to glucosylceramide, miglustat was found to perform a chaperone function that increased the activity of β-glucosidase61, which inspired a search for novel analogues with similar activity62. Although this approach is promising, no examples currently exist of drugs that have been repurposed in this way to treat cardiovascular diseases.
Experimental methods of drug repurposing
Phenotypic screening
Historically, most drugs were discovered based on their effects on phenotype; however, in the 1970s, target-based drug discovery driven by the systematic screening of large chemical libraries became the primary source of identifying new drug candidates63. Nevertheless, phenotypic drug discovery is again being commonly used in drug discovery but, unlike historical discovery based on observation, modern phenotypic discovery involves systematic screening of molecules for disease-modifying activity64. Phenotypic discovery has been shown to be a reliable method for the discovery and development of first-in-class drugs65 and is arguably more effective than target-based drug discovery in identifying compounds that succeed in clinical development66,67. Given that phenotypic drug discovery is a target-agnostic approach, phenotypic screening of approved and investigational drugs is highly appropriate for the discovery of drug repurposing candidates.
One of the more advanced compounds repurposed for a cardiovascular indication that originated from in vitro phenotypic screening is REC-994, which is now in clinical development for the treatment of cerebral cavernous malformations. These malformations are collections of enlarged and irregular capillaries in the brain that alter blood flow and cause severe symptoms, such as seizures, vision and hearing loss, paralysis and haemorrhagic stroke. Initiated by a phenotypic primary assay that evaluated the normalization of cellular features detected by image analysis in human microvascular endothelial cells deficient in cerebral cavernous malformations 2 protein (CCM2; encoded by CCM2, one of at least three genes associated with these malformations), active compounds were then triaged by functional studies in the same in vitro model and subsequently in inducible Ccm2-knockout mice68. Using the open-source Cell Painting algorithm as an assay, the primary screen quantified the reversal of machine-learned features indicative of CCM2 loss of function in the endothelial cells, yielding two hits (out of 2,100 known drugs screened) — cholecalciferol (vitamin D3) and tempol (designated REC-994) — that reduced lesion burden in the Ccm2-knockout mice68.
The target-agnostic nature of phenotypic screening can be a valuable entry point into the discovery of novel mechanisms. Illustrative of this concept is the programme to develop small-molecule inhibitors of proprotein convertase subtilisin/kexin type 9 (PCSK9) for the treatment of dyslipidaemia. PCSK9 inhibitors were inspired by the realization that individuals carrying loss-of-function variants in PCSK9 have low serum levels of LDL cholesterol and unusually good cardiovascular health69. A phenotypic screen measuring PCSK9 secretion led to the identification of a compound that was refined to yield potent compounds that bind to the human 80S ribosome and stall the translation of PCSK9 mRNA. Remarkably, the effect is selective for PCSK9 and a few other proteins in the cell70. Small-molecule-directed, selective ribosome stalling was completely unanticipated and could potentially be directed to other therapeutic target proteins.
The major shortcoming of using phenotypic assays for drug repurposing is that the targets and mechanisms of action responsible for the new indication might be different from those that are responsible for the known effects of a drug. Small molecules are never absolutely selective, making it a challenge to define targets and mechanisms of action de novo. Target information is useful because it can help to identify safety risks (less of a concern for repurposed drugs than for new chemical entities) and inform the development of tests to monitor efficacy during clinical development63. Although lack of knowledge about the direct targets is not an absolute impediment to advancing a compound, it can nonetheless be perceived as a serious obstacle to further development in pharmaceutical companies and by regulatory authorities63. Various strategies for target identification are available, including affinity chromatography and mass spectrometry, as reviewed previously63. Particularly powerful are mass spectrometry approaches that quantify chemical binding based on altered thermostability (for example, the cellular thermal shift assay)71. Given that the target information is useful for clinical advancement and might also provide new mechanistic insights, target identification is a logical adjunct to drug repurposing studies.
iPSC models of heart disease
Numerous induced pluripotent stem cell (iPSC) models have been developed for drug screening because they reproduce clinical features of genetic, electrophysiological, myopathic and metabolic disorders as well as clinically relevant drug responses72,73. For example, treatment of iPSC-derived cardiomyocytes (iPSC-CMs) with known drugs evokes the expected physiological effects on cardiac action potentials, intracellular calcium transients and cellular contractility72,74.
Given that iPSCs reflect the genetics of the individual donor, they offer an unprecedented opportunity to focus drug screening on congenital heart disease and have been widely used to explore therapeutic strategies (Fig. 2). For example, iPSC-CMs derived from patients with idiopathic hypertrophic cardiomyopathy revealed that Pyr3, an inhibitor of the short transient receptor potential channel 3 (TRPC3), attenuates the upregulated diastolic calcium levels and decreases cardiomyocyte hypertrophy across diverse aetiologies of hypertrophic cardiomyopathy75. Using patient-derived and CRISPR-mediated gene-edited iPSC-CM models, we found that small-molecule activators of the unfolded protein response and RBM20 transcriptional upregulation can normalize the contractile deficits that are characteristic of dilated cardiomyopathy associated with variants in PLN and RBM20, respectively76,77. iPSC-derived cardiac cells are well-suited to modelling inherited cardiovascular diseases because these diseases often have a cell-autonomous aetiology, are early-onset (which facilitates modelling in monocultures) and are also often monogenic (enabling the generation of isogenic control lines by gene editing)73. Diseases with these traits tend to be rare, so the use of iPSC models to investigate drug repositioning could help to address the deficit in new therapeutics for rare cardiovascular diseases.
a | Induced pluripotent stem cells (iPSCs), derived from healthy donors or donors carrying genetic variants that predispose them to cardiovascular disease, are directed to differentiate into cardiovascular cell types with the use of protocols that mimic mechanisms of early development. Depending on the presence of gene variants and/or the culture conditions, the differentiated derivative cells can display phenotypes that are consistent with a genetic or acquired disease. b | These iPSC models are used to screen drug repurposing candidates, resulting in the identification of drugs for preclinical and clinical testing. c | If the drug candidates have remediable limitations for their new indication, they can be refined through iterative cycles of medicinal chemical optimization and in vitro testing, aided by iPSC-based assays.
iPSC-based screening has not yet resulted in a repurposed or re-engineered drug that has progressed to clinical use in cardiovascular disease, although iPSC modelling has led to clinical testing of repurposed drugs for amyotrophic lateral sclerosis78,79,80. A few examples exist of iPSC-CMs being used for large-scale discovery re-engineering. In one instance, researchers developed an iPSC-CM model of diabetic cardiomyopathy and identified molecules that blunted disease features and revealed aberrant protein homeostasis as a potential disease mechanism and therapeutic target81. In a second example, investigators developed an iPSC-CM model of oxidative stress and used the system to optimize novel inhibitors of mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4) that were subsequently shown to reduce ischaemia–reperfusion injury in mice82. A third example was the use of patient-derived iPSC-CMs to re-engineer a small-molecule antiarrhythmic drug83,84, as described in the re-engineering section below.
A limitation of iPSC-CMs is that they are physiologically immature and resemble fetal rather than adult cardiomyocytes. Given that iPSC-CMs lack certain electrophysiological, structural and mechanical properties that arise postnatally, their use in drug discovery could inadvertently lead to the identification of drug actions relevant to the fetal rather than the adult heart. Efforts to overcome physiological immaturity include the use of electrical stimulation, patterned surfaces and culture media supplements73. Another limitation is that most systems for drug screening favour throughput over realism and, therefore, tend to be 2D sheets of confluent cells (such as cardiomyocytes). Although extremely useful, these systems are limited in their capacity to mimic tissue-level or organ-level physiological functions involving multiple cell types. Substantial progress has been made in developing 3D tissue and microphysiological systems that can be used to detect drug and genetic effects85,86.
Zebrafish as screening tools
The development of zebrafish models of human cardiovascular disease and their validation by recapitulating expected pharmacological responses laid the groundwork for their popularity as a screening tool87. The advantage of using zebrafish is that they model the complexity of disease in the context of the whole organism, but can still be used for moderate-throughput screening, exemplified by chemical screens that identified compounds that could rescue developmental cardiac defects88,89. Subsequent studies identified disease-modifying compounds that shorten the ventricular action potential in zebrafish models of long QT syndrome type 2 (ref.90) or inhibit ocular angiogenesis91. Although most chemical screens have used larval zebrafish, transgenic lines carrying fluorescent sensors have made it possible to screen for drugs that modulate cardiac function in juvenile and adult zebrafish92. Several drugs identified for repurposing as a result of screening in zebrafish entered clinical development in the early 2010s, including compounds to improve the engraftment of donor haematopoietic stem cells in patients with leukaemia93,94 and the use of clemizole in patients with Dravet syndrome95,96, although none has progressed to clinical approval.
Trametinib, a mitogen-activated protein kinase kinase (MEK) inhibitor, was found to rescue the lymphatic phenotype in a larval zebrafish model of a gain-of-function variant in ARAF identified in a patient with advanced anomalous lymphatic disease that was unresponsive to conventional sirolimus therapy97. Treatment of the patient with trametinib for 1 year yielded nearly complete regression of lymphoedema, allowing the child to return to school97. Subsequently, trametinib was used to treat lymphatic anomaly in a second patient with a variant in SOS1, which encodes a protein acting on the MEK–MAPK pathway98. Of note, the effect of treatment seemed to be comprehensive in both patients, with systemic remodelling of the entire central lymphatic system. No information on toxicities was reported, and additional studies are warranted to explore durability, reversibility and adverse effects of these drugs.
Similar zebrafish models of arteriovenous malformations caused by variants in BRAF or MAP2K1 (which act upstream in the RAF–RAS–MAPK cascade) provided proof-of-concept that vemurafenib, a BRAF inhibitor, could improve blood flow, mimicking results seen in mouse models99. Interestingly, trametinib is now under evaluation in a phase II clinical trial for extracranial arteriovenous malformations100, with completion of the trial expected in 2022. Although the pace of drug repurposing and new-entity discovery using zebrafish screening seems to have lost ground relative to the use of iPSCs, zebrafish occupy a unique niche as a ‘screenable’ vertebrate.
Drug re-engineering
Some of the same computational and experimental tools that can be used to repurpose drugs can also be applied to refine the chemical composition of existing drugs to improve their pharmacological or therapeutic properties. Drug refinement is likely to become a valuable adjunct to drug repurposing, for example when an existing drug has been repurposed because of a molecular mechanism of action or polypharmacology that is distinct from that for which it was originally developed. In this case, the newly repurposed ‘hit’ might have limitations (such as low potency) that limit its usefulness for the new indication. If the previously unknown targets that mediate the new function can be identified, a conventional, target-driven approach to hit optimization could be used in part to drive the medicinal chemistry refinement of the compound. Conversely, phenotypic assays might be useful to guide refinement when the candidate functions in its new context through unknown or complicated mechanisms. For example, iPSC-based models of cardiometabolic disease have been used in high-throughput studies to refine new chemical entities81,82. However, detailed structure–activity relationship studies are necessary because subtle chemical modifications can profoundly alter biological activity. Numerous strategies can be used to help to ensure the in vivo efficacy of compounds identified by in vitro phenotypic screening64. Relevant to refining repurposed compounds, these approaches include assays that track molecular signatures (such as the transcriptome-matching studies described above) to verify engagement of disease-modifying mechanisms to improve the likelihood of in vivo efficacy.
Evidence suggests that iPSC-based phenotypic models of heart disease have the potential to drive re-engineering for cardiovascular indications (Fig. 2). In the case of the MAP4K4 inhibitors for myocardial ischaemia discussed above, the researchers used the iPSC-CM model to select potent analogue compounds on the basis of increased survival after exposure to oxidative stress82. In addition, we described the use of iPSC-CMs derived from patients with long QT syndrome type 3 (with variants in SCN5A, encoding the Nav1.5 channel) to guide the medicinal chemistry refinement of the antiarrhythmic drug mexiletine83,84. By improving the potency and selectivity for blocking the cardiac late sodium current, the new analogues might be useful to block the pathological rise in intracellular sodium and calcium levels and the activation of calcium/calmodulin-dependent protein kinase II that promote ventricular tachycardia and ventricular fibrillation in these patients101.
A re-engineered drug or a repurposed drug with regulatory approval for its original indication will need to demonstrate an acceptable benefit–risk ratio in the new indication. New regulatory approval will be required, usually necessitating a clinical study for the new indication.
Future directions
Modern drug repurposing takes advantage of electronic health records, genomics and iPSC models that recapitulate disease. Each of these resources embodies individual patient genetics and disease to a highly personalized degree and, as such, the use of these technologies in the discovery phase has two major implications for repurposed drugs. The first is that the highly detailed and personal knowledge of the patient population that the repurposed drug is intended to benefit can be consolidated and used to aid clinicians in precisely prescribing the drug. Realizing this benefit might require the development of diagnostic tools that can discriminate between patients who might benefit and patients who might not benefit. The second implication is that drug repurposing via these patient-focused technologies might have a disproportionate effect on rare cardiovascular diseases, for which the financial returns on investment are often insufficient to support drug development from concept to bedside.
Repurposed drugs might need to be optimized for their new indications, and we discussed the use of iPSCs and other phenotypic strategies to guide medicinal chemistry re-engineering. In principle, the same cardiovascular assays could be applied to re-engineer existing drugs to decrease any cardiovascular liabilities. Oncology therapeutics are one such opportunity. For example, vascular and cardiomyopathic effects of oncology therapeutics have been modelled in iPSC-CM and zebrafish models102,103, establishing the groundwork to explore the chemical basis for the cardiotoxic effects of oncology therapeutics.
With ever-increasing computational power, the efforts described above that match drugs to new indications could possibly be extended to predict the use of drugs to treat subgroups of patients within already approved indications or to stratify patients for recruitment into clinical trials104. However, time will tell if either of these possibilities will be realized.
Finally, we have only touched on the technical challenges of defining disease signatures from omics and clinical data or from complex phenotypic screening. Increasing attention is being given to the use of machine learning to define signatures of disease105. Although the greatest effect of artificial intelligence has been in disease diagnosis and gleaning information from electronic health records, machine-learned features of highly complex datasets have the potential to associate drug repurposing candidates or their effects with complex disease phenotypes that are apparent in histopathology or electrophysiology datasets, which could be applied to drug screening and repurposing. The use of machine learning in drug discovery has been reviewed previously106,107.
Conclusions
Systematic drug repurposing has created a new landscape of opportunities. To date, however, few clinical successes have been reported in cardiovascular medicine. To some extent, this situation reflects the recent emergence of systematic drug repurposing, but it also suggests that substantial barriers might have to be overcome. Some are inherent to the discovery of any new drug, such as failure to demonstrate an acceptable benefit–risk ratio in clinical trials, although toxicity is less likely to be a problem for repurposed drugs. Other barriers are more specific to drug repurposing and include the protection of intellectual property, which diminishes the financial incentive for repurposing108,109. Compounding this issue is that the powerful iPSC (and zebrafish) technologies are most readily suited to modelling monogenic diseases. Given that monogenic diseases tend to be rare, the small market size for the eventual therapeutic agent can be a disincentive. To address intellectual property barriers, schemes that allow second medical use patents have been introduced in some markets; however, this remedy might not sufficiently incentivize repurposing because of the need to demonstrate non-obviousness and avoid potential competition from pre-existing generics108 as well as market concerns in the case of rare diseases. Therefore, reforms that incentivize drug repurposing programmes and create an adequate return on investment that is sufficient to motivate the effort might accelerate systematic drug repurposing and re-engineering. Specific recommendations have been made to realize the potential of drug repurposing109 (Box 2).
In summary, we have entered a new era of deliberate drug repurposing driven by the convergence of computational and experimental technologies. The computational tools build on the ability to generate and manipulate large multiparametric datasets describing molecular and phenotypic features of disease and similar datasets of drug effects, while the experimental tools comprise realistic disease models capable of high-throughput testing of drug repurposing candidates. From the perspective of precision medicine, these tools make it possible to match the mechanisms of action of an existing drug to the molecular dysfunctions that cause disease and, if needed, optimize the drug on the basis of disease-modifying effects. With support and incentivization, the new computational and experimental technologies could increase the effectiveness of cardiovascular drug discovery and the range of diseases that can be treated.
References
Estruch, R., Ruilope, L. M. & Cosentino, F. The year in cardiovascular medicine 2020: epidemiology and prevention. Eur. Heart J. 42, 813–821 (2021).
Mullard, A. 2019 FDA drug approvals. Nat. Rev. Drug Discov. 19, 79–84 (2020).
Moore, T. J., Zhang, H., Anderson, G. & Alexander, G. C. Estimated costs of pivotal trials for novel therapeutic agents approved by the US Food and Drug Administration, 2015–2016. JAMA Intern. Med. 178, 1451–1457 (2018).
Fordyce, C. B. et al. Cardiovascular drug development: is it dead or just hibernating? J. Am. Coll. Cardiol. 65, 1567–1582 (2015).
Kort, E. & Jovinge, S. Drug repurposing: claiming the full benefit from drug development. Curr. Cardiol. Rep. 23, 62 (2021).
Lee, A. F., MacFadyen, R. J. & Struthers, A. D. Neurohormonal reactivation in heart failure patients on chronic ACE inhibitor therapy: a longitudinal study. Eur. J. Heart Fail. 1, 401–406 (1999).
Lacaze, P. et al. Genetic variants associated with inherited cardiovascular disorders among 13,131 asymptomatic older adults of European descent. NPJ Genom. Med. 6, 51 (2021).
Abdulrahim, J. W. et al. Identification of undetected monogenic cardiovascular disorders. J. Am. Coll. Cardiol. 76, 797–808 (2020).
Gelosa, P., Castiglioni, L., Camera, M. & Sironi, L. Repurposing of drugs approved for cardiovascular diseases: opportunity or mirage? Biochem. Pharmacol. 177, 113895 (2020).
MarketWatch. Global minoxidil market 2022–2028: worldwide industry growing at a CAGR of 4.5% and expected to reach USD 1335.6 million. https://www.marketwatch.com/press-release/global-minoxidil-market-2022-2028-worldwide-industry-growing-at-a-cagr-of-45-and-expected-to-reach-usd-13356-million-2022-02-25 (2022).
Montinari, M. R., Minelli, S. & De Caterina, R. The first 3500 years of aspirin history from its roots–a concise summary. Vasc. Pharmacol. 113, 1–8 (2019).
Kim, J. et al. Impact of blockade of histamine H2 receptors on chronic heart failure revealed by retrospective and prospective randomized studies. J. Am. Coll. Cardiol. 48, 1378–1384 (2006).
Leary, P. J. et al. Histamine H2 receptor antagonists, left ventricular morphology, and heart failure risk: the MESA study. J. Am. Coll. Cardiol. 67, 1544–1552 (2016).
Zhang, J. et al. Cardioprotective effect of histamine H2 antagonists in congestive heart failure: a systematic review and meta-analysis. Medicine 97, e0409 (2018).
Banach, M. & Penson, P. E. Colchicine and cardiovascular outcomes: a critical appraisal of recent studies. Curr. Atheroscler. Rep. 23, 32 (2021).
Tien, Y. Y., Huang, H. K., Shih, M. C. & Tu, Y. K. Drug repurposing? Cardiovascular effect of colchicine on patients with coronary artery disease: a systematic review and meta-analysis. J. Cardiol. 77, 576–582 (2021).
Nidorf, S. M. & Thompson, P. L. Why colchicine should be considered for secondary prevention of atherosclerosis: an overview. Clin. Ther. 41, 41–48 (2019).
Kalkhoran, S. B. et al. Hydralazine protects the heart against acute ischemia/reperfusion injury by inhibiting Drp1-mediated mitochondrial fission. Cardiovasc. Res. 118, 282–294 (2022).
Porto, R. et al. Antiparasitic properties of cardiovascular agents against human intravascular parasite Schistosoma mansoni. Pharmaceuticals 14, 686 (2021).
Jiang, W., Hu, J. W., He, X. R., Jin, W. L. & He, X. Y. Statins: a repurposed drug to fight cancer. J. Exp. Clin. Cancer Res. 40, 241 (2021).
McGowan, N. M., Nichols, M., Bilderbeck, A. C., Goodwin, G. M. & Saunders, K. E. A. Blood pressure in bipolar disorder: evidence of elevated pulse pressure and associations between mean pressure and mood instability. Int. J. Bipolar Disord. 9, 5 (2021).
Umbarkar, P. et al. Repurposing nintedanib for pathological cardiac remodeling and dysfunction. Pharmacol. Res. 169, 105605 (2021).
Huang, Y. et al. Rapid repurposing of novel combination drugs for the treatment of heart failure via a computationally guided network screening approach. J. Chem. Inf. Model https://doi.org/10.1021/acs.jcim.1c00132 (2021).
Chapman, W. W. et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J. Am. Med. Inf. Assoc. 18, 540–543 (2011).
Schubert, M., Hansen, S., Leefmann, J. & Guan, K. Repurposing antidiabetic drugs for cardiovascular disease. Front. Physiol. 11, 568632 (2020).
Mordor Intelligence. Sodium-dependent glucose co-transporter 2 (SGLT2) market–growth, trends, COVID-19 impact, and forecasts (2022–2027). https://www.mordorintelligence.com/industry-reports/sglt-2-market (2022).
Zannad, F. et al. SGLT2 inhibitors in patients with heart failure with reduced ejection fraction: a meta-analysis of the EMPEROR-Reduced and DAPA-HF trials. Lancet 396, 819–829 (2020).
Nassif, M. E. et al. The SGLT2 inhibitor dapagliflozin in heart failure with preserved ejection fraction: a multicenter randomized trial. Nat. Med. 27, 1954–1960 (2021).
Weeber, M., Kors, J. A. & Mons, B. Online tools to support literature-based discovery in the life sciences. Brief. Bioinform 6, 277–286 (2005).
Weeber, M. et al. Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide. J. Am. Med. Inf. Assoc. 10, 252–259 (2003).
Milazzo, L. et al. Thalidomide in the treatment of chronic hepatitis C unresponsive to alfa-interferon and ribavirin. Am. J. Gastroenterol. 101, 399–402 (2006).
Liu, P. P. et al. Aspirin alleviates cardiac fibrosis in mice by inhibiting autophagy. Acta Pharmacol. Sin. 38, 488–497 (2017).
Hoffmann, M. et al. Repurposing mesalazine against cardiac fibrosis in vitro. Naunyn Schmiedebergs Arch. Pharmacol. 394, 533–543 (2021).
Campillos, M., Kuhn, M., Gavin, A. C., Jensen, L. J. & Bork, P. Drug target identification using side-effect similarity. Science 321, 263–266 (2008).
Xu, H. et al. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J. Am. Med. Inf. Assoc. 22, 179–191 (2015).
Gandini, S. et al. Metformin and cancer risk and mortality: a systematic review and meta-analysis taking into account biases and confounders. Cancer Prev. Res. 7, 867–885 (2014).
Yu, H. et al. The potential effect of metformin on cancer: an umbrella review. Front. Endocrinol. 10, 617 (2019).
Bragagnoli, A. C. et al. Metformin plus lrinotecan in patients with refractory colorectal cancer: a phase 2 clinical trial. Br. J. Cancer 124, 1072–1078 (2021).
Brown, J. R. et al. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight 5, e133247 (2020).
Lau, A. & So, H. C. Turning genome-wide association study findings into opportunities for drug repositioning. Comput. Struct. Biotechnol. J. 18, 1639–1650 (2020).
Teslovich, T. M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Law, M. R., Wald, N. J. & Rudnicka, A. R. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. BMJ 326, 1423 (2003).
Sanseau, P. et al. Use of genome-wide association studies for drug repositioning. Nat. Biotechnol. 30, 317–320 (2012).
Grover, M. P. et al. Novel therapeutics for coronary artery disease from genome-wide association study data. BMC Med. Genomics 8, S1 (2015).
Tragante, V. et al. Druggability of coronary artery disease risk loci. Circ. Genom. Precis. Med. 11, e001977 (2018).
Zaghlool, S. B. et al. Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat. Commun. 12, 1279 (2021).
Folkersen, L. et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat. Metab. 2, 1135–1148 (2020).
Allen, F. H., Kennard, O., Motherwell, W. D. S., Town, W. G. & Watson, D. G. Cambridge Crystallographic Data Centre. II. Structural data file. J. Chem. Doc. 13, 119–123 (1973).
May, E. E., Leitao, A., Tropsha, A. & Oprea, T. I. A systems chemical biology study of malate synthase and isocitrate lyase inhibition in Mycobacterium tuberculosis during active and NRP growth. Comput. Biol. Chem. 47, 167–180 (2013).
Keiser, M. J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009).
Zheng, C. et al. Large-scale direct targeting for drug repositioning and discovery. Sci. Rep. 5, 11970 (2015).
Ribaudo, G. et al. A computational approach to drug repurposing against SARS-CoV-2 RNA dependent RNA polymerase (RdRp). J. Biomol. Struct. Dyn. 40, 1101–1108 (2022).
Toro-Domínguez, D., Alarcón-Riquelme, M. E. & Carmona-Sáez, P. in In Silico Drug Design Ch. 11 (ed. Roy, K.) 303–327 (Elsevier, 2019).
Dudley, J. T. et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 3, 96ra76 (2011).
Crockett, S. D., Schectman, R., Sturmer, T. & Kappelman, M. D. Topiramate use does not reduce flares of inflammatory bowel disease. Dig. Dis. Sci. 59, 1535–1543 (2014).
Kort, E. J. & Jovinge, S. Streamlined analysis of LINCS L1000 data with the slinky package for R. Bioinformatics 35, 3176–3177 (2019).
Kort, E. J., Sayed, N., Liu, C., Wu, J. C. & Jovinge, S. Toxicogenomic identification of repositioned therapy for a monogenic disease. bioRXiv https://doi.org/10.1101/748863 (2019).
Van Vleet, T. R., Liguori, M. J., Lynch, J. J. 3rd, Rao, M. & Warder, S. Screening strategies and methods for better off-target liability prediction and identification of small-molecule pharmaceuticals. SLAS Discov. 24, 1–24 (2019).
Gfeller, D. et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 42, W32–W38 (2014).
Lee, Y. H., Choi, H., Park, S., Lee, B. & Yi, G. S. Drug repositioning for enzyme modulator based on human metabolite-likeness. BMC Bioinforma. 18, 226 (2017).
Sawkar, A. R. et al. Chemical chaperones increase the cellular activity of N370S β-glucosidase: a therapeutic strategy for Gaucher disease. Proc. Natl Acad. Sci. USA 99, 15428–15433 (2002).
Sanchez-Olle, G. et al. Promising results of the chaperone effect caused by imino sugars and aminocyclitol derivatives on mutant glucocerebrosidases causing Gaucher disease. Blood Cell Mol. Dis. 42, 159–166 (2009).
Moffat, J. G., Vincent, F., Lee, J. A., Eder, J. & Prunotto, M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug. Discov. 16, 531–543 (2017).
Vincent, F. et al. Phenotypic drug discovery: recent successes, lessons learned and new directions. Nat. Rev. Drug Discov. https://doi.org/10.1038/s41573-022-00472-w (2022).
Swinney, D. C. & Lee, J. A. Recent advances in phenotypic drug discovery. F1000Res 9, 944 (2020).
Swinney, D. C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug. Discov. 10, 507–519 (2011).
Scannell, J. W. & Bosley, J. When quality beats quantity: decision theory, drug discovery, and the reproducibility crisis. PLoS ONE 11, e0147215 (2016).
Gibson, C. C. et al. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation 131, 289–299 (2015).
Cohen, J. C., Boerwinkle, E., Mosley, T. H. Jr. & Hobbs, H. H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 (2006).
Lintner, N. G. et al. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol. 15, e2001882 (2017).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Del Alamo, J. C. et al. High throughput physiological screening of iPSC-derived cardiomyocytes for drug development. Biochim. Biophys. Acta 1863, 1717–1727 (2016).
Hnatiuk, A. P., Briganti, F., Staudt, D. W. & Mercola, M. Human iPSC modeling of heart disease for drug development. Cell Chem. Biol. 28, 271–282 (2021).
Paik, D. T., Chandy, M. & Wu, J. C. Patient and disease-specific induced pluripotent stem cells for discovery of personalized cardiovascular drugs and therapeutics. Pharmacol. Rev. 72, 320–342 (2020).
Sakai, T. et al. Phenotypic screening using patient-derived induced pluripotent stem cells identified Pyr3 as a candidate compound for the treatment of infantile hypertrophic cardiomyopathy. Int. Heart J. 59, 1096–1105 (2018).
Briganti, F. et al. iPSC modeling of RBM20-deficient DCM identifies upregulation of RBM20 as a therapeutic strategy. Cell Rep. 32, 108117 (2020).
Feyen, D. A. M. et al. The unfolded protein response as a compensatory mechanism and potential therapeutic target in PLN R14del cardiomyopathy. Circulation 144, 382–392 (2021).
Wainger, B. J. et al. Effect of ezogabine on cortical and spinal motor neuron excitability in amyotrophic lateral sclerosis: a randomized clinical trial. JAMA Neurol. 78, 186–196 (2021).
Morimoto, S. et al. Ropinirole hydrochloride remedy for amyotrophic lateral sclerosis–protocol for a randomized, double-blind, placebo-controlled, single-center, and open-label continuation phase I/IIa clinical trial (ROPALS trial). Regen. Ther. 11, 143–166 (2019).
Imamura, K. et al. Induced pluripotent stem cell-based drug repurposing for amyotrophic lateral sclerosis medicine (iDReAM) study: protocol for a phase I dose escalation study of bosutinib for amyotrophic lateral sclerosis patients. BMJ Open. 9, e033131 (2019).
Drawnel, F. M. et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 9, 810–821 (2014).
Fiedler, L. R. et al. MAP4K4 inhibition promotes survival of human stem cell-derived cardiomyocytes and reduces infarct size in vivo. Cell Stem Cell 24, 579–591.e12 (2019).
McKeithan, W. L. et al. Reengineering an antiarrhythmic drug using patient hiPSC cardiomyocytes to improve therapeutic potential and reduce toxicity. Cell Stem Cell 27, 813–821.e6 (2020).
Cashman, J. R. et al. Antiarrhythmic hit to lead refinement in a dish using patient-derived iPSC cardiomyocytes. J. Med. Chem. 64, 5384–5403 (2021).
Mathur, A. et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 5, 8883 (2015).
Bliley, J. M. et al. Dynamic loading of human engineered heart tissue enhances contractile function and drives a desmosome-linked disease phenotype. Sci. Transl. Med. 13, eabd1817 (2021).
Pott, A., Rottbauer, W. & Just, S. Streamlining drug discovery assays for cardiovascular disease using zebrafish. Expert Opin. Drug Discov. 15, 27–37 (2020).
Peterson, R. T. et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 22, 595–599 (2004).
Hong, C. C., Peterson, Q. P., Hong, J. Y. & Peterson, R. T. Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr. Biol. 16, 1366–1372 (2006).
Peal, D. S. et al. Novel chemical suppressors of long QT syndrome identified by an in vivo functional screen. Circulation 123, 23–30 (2011).
Reynolds, A. L. et al. Phenotype-based discovery of 2-[(E)-2-(quinolin-2-yl)vinyl]phenol as a novel regulator of ocular angiogenesis. J. Biol. Chem. 291, 7242–7255 (2016).
van Opbergen, C. J. M. et al. Optogenetic sensors in the zebrafish heart: a novel in vivo electrophysiological tool to study cardiac arrhythmogenesis. Theranostics 8, 4750–4764 (2018).
Cutler, C. et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood 122, 3074–3081 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT00890500 (2013).
Baraban, S. C., Dinday, M. T. & Hortopan, G. A. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat. Commun. 4, 2410 (2013).
US National Library of Medicine. ClinicalTrials.gov http://www.clinicaltrials.gov/ct2/show/NCT04462770 (2022).
Li, D. et al. ARAF recurrent mutation causes central conducting lymphatic anomaly treatable with a MEK inhibitor. Nat. Med. 25, 1116–1122 (2019).
Dori, Y. et al. Severe lymphatic disorder resolved with MEK inhibition in a patient with Noonan syndrome and SOS1 mutation. Pediatrics 146, e20200167 (2020).
Al-Olabi, L. et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest. 128, 1496–1508 (2018).
US National Library of Medicine. ClinicalTrials.gov http://www.clinicaltrials.gov/ct2/show/NCT04258046 (2022).
Karagueuzian, H. S., Pezhouman, A., Angelini, M. & Olcese, R. Enhanced late Na and Ca currents as effective antiarrhythmic drug targets. Front. Pharmacol. 8, 36 (2017).
Liu, Y. et al. Visnagin protects against doxorubicin-induced cardiomyopathy through modulation of mitochondrial malate dehydrogenase. Sci. Transl. Med. 6, 266ra170 (2014).
Sharma, A. et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 9, eaaf2584 (2017).
Inoue, H., Nagata, N., Kurokawa, H. & Yamanaka, S. iPS cells: a game changer for future medicine. EMBO J. 33, 409–417 (2014).
Schuhmacher, A., Gatto, A., Hinder, M., Kuss, M. & Gassmann, O. The upside of being a digital pharma player. Drug Discov. Today 25, 1569–1574 (2020).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Schneider, P. et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 19, 353–364 (2020).
Breckenridge, A. & Jacob, R. Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. 18, 1–2 (2019).
Pushpakom, S. et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18, 41–58 (2019).
Guo, C. G. et al. Aspirin use and risk of colorectal cancer among older adults. JAMA Oncol. 7, 428–435 (2021).
Clemow, D. B. & Bushe, C. J. Atomoxetine in patients with ADHD: a clinical and pharmacological review of the onset, trajectory, duration of response and implications for patients. J. Psychopharmacol. 29, 1221–1230 (2015).
McMahon, C. G. Dapoxetine: a new option in the medical management of premature ejaculation. Ther. Adv. Urol. 4, 233–251 (2012).
Maund, E., Guski, L. S. & Gotzsche, P. C. Considering benefits and harms of duloxetine for treatment of stress urinary incontinence: a meta-analysis of clinical study reports. CMAJ 189, E194–E203 (2017).
Pelletier, D. & Hafler, D. A. Fingolimod for multiple sclerosis. N. Engl. J. Med. 366, 339–347 (2012).
Shirley, M. Ketoconazole in Cushing’s syndrome: a profile of its use. Drugs Ther. Persp 37, 55–64 (2021).
Vogel, V. G. et al. Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: the NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. JAMA 295, 2727–2741 (2006).
Buch, M. H. et al. Updated consensus statement on the use of rituximab in patients with rheumatoid arthritis. Ann. Rheum. Dis. 70, 909–920 (2011).
Mikulic, M. Worldwide revenue of Pfizer’s Viagra from 2003 to 2019. Statista https://www.statista.com/statistics/264827/pfizers-worldwide-viagra-revenue-since-2003/ (2021).
Singhal, S. et al. Antitumor activity of thalidomide in refractory multiple myeloma. N. Engl. J. Med. 341, 1565–1571 (1999).
Kramer, C. K. et al. Efficacy and safety of topiramate on weight loss: a meta-analysis of randomized controlled trials. Obes. Rev. 12, e338–e347 (2011).
Yarchoan, R. et al. Administration of 3′-azido-3′-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. Lancet 1, 575–580 (1986).
Ridker, P. M. et al. Effects of interleukin-1β inhibition with canakinumab on hemoglobin A1c, lipids, C-reactive protein, interleukin-6, and fibrinogen: a phase IIb randomized, placebo-controlled trial. Circulation 126, 2739–2748 (2012).
Everett, B. M. et al. Anti-inflammatory therapy with canakinumab for the prevention of hospitalization for heart failure. Circulation 139, 1289–1299 (2019).
Crittenden, D. B. et al. Colchicine use is associated with decreased prevalence of myocardial infarction in patients with gout. J. Rheumatol. 39, 1458–1464 (2012).
Tardif, J. C. et al. Efficacy and safety of low-dose colchicine after myocardial infarction. N. Engl. J. Med. 381, 2497–2505 (2019).
Sato, K. et al. The effect of donepezil treatment on cardiovascular mortality. Clin. Pharmacol. Ther. 88, 335–338 (2010).
Lonborg, J. et al. Exenatide reduces reperfusion injury in patients with ST-segment elevation myocardial infarction. Eur. Heart J. 33, 1491–1499 (2012).
Ratner, R. et al. Cardiovascular safety of exenatide BID: an integrated analysis from controlled clinical trials in participants with type 2 diabetes. Cardiovasc. Diabetol. 10, 22 (2011).
Habashi, J. P. et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science 312, 117–121 (2006).
van Andel, M. M. et al. Long-term clinical outcomes of losartan in patients with Marfan syndrome: follow-up of the multicentre randomized controlled COMPARE trial. Eur. Heart J. 41, 4181–4187 (2020).
Hwang, I. C. et al. Pulmonary hemodynamics and effects of phosphodiesterase type 5 inhibition in heart failure: a meta-analysis of randomized trials. BMC Cardiovas Disord. 17, 150 (2017).
Andersson, D. P. et al. Association between treatment for erectile dysfunction and death or cardiovascular outcomes after myocardial infarction. Heart 103, 1264–1270 (2017).
Kim, S. C. et al. Cardiovascular safety of tocilizumab versus tumor necrosis factor inhibitors in patients with rheumatoid arthritis: a multi-database cohort study. Arthritis Rheumatol. 69, 1154–1164 (2017).
Broch, K. et al. Randomized trial of interleukin-6 receptor inhibition in patients with acute ST-segment elevation myocardial infarction. J. Am. Coll. Cardiol. 77, 1845–1855 (2021).
Acknowledgements
M.A. is a recipient of a Heart Rhythm Society postdoctoral fellowship. The authors gratefully acknowledge grant support from the Richard and Helen DeVos Foundation (2014-Jovinge) to E.J.K. and S.J. and from the NIH (R01HL138539, R01HL152055 and P01HL141084) and the Fondation Leducq (CUREPLaN) to M.M.
Author information
Authors and Affiliations
Contributions
M.A., S.J. and M.M. researched data for the article. All the authors contributed substantially to discussion of the content and wrote the article. M.A., S.J. and M.M. reviewed and/or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Cardiology thanks Kaomei Guan, who co-reviewed with Mario Schubert; Jean-Pierre Valentin; Xi Yang; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CSD: https://www.ccdc.cam.ac.uk/solutions/csd-core/components/csd/
EudraVigilance: https://www.ema.europa.eu/en/human-regulatory/research-development/pharmacovigilance/eudravigilance
FAERS: https://open.fda.gov/data/faers/
LINCS: https://lincsproject.org/
SwissTargetPrediction: http://www.swisstargetprediction.ch/
UMLS: https://www.nlm.nih.gov/research/umls/index.html
VigiBase: https://who-umc.org/vigibase/
Rights and permissions
About this article
Cite this article
Abdelsayed, M., Kort, E.J., Jovinge, S. et al. Repurposing drugs to treat cardiovascular disease in the era of precision medicine. Nat Rev Cardiol 19, 751–764 (2022). https://doi.org/10.1038/s41569-022-00717-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-022-00717-6
This article is cited by
-
Testing the predictive power of reverse screening to infer drug targets, with the help of machine learning
Communications Chemistry (2024)
-
Repurposing of IL 33/ST2 Modulating Drugs as a Cardioprotective Agent: A Promising Approach
Journal of Pharmaceutical Innovation (2024)
-
Systems immunology approach for targeting inflammation in atherosclerosis
Nature Cardiovascular Research (2023)
-
Ruxolitinib is a CaMKII inhibitor that can be repurposed to prevent arrhythmias
Nature Reviews Cardiology (2023)
-
Systems immunology-based drug repurposing framework to target inflammation in atherosclerosis
Nature Cardiovascular Research (2023)