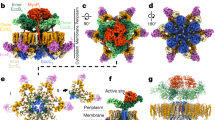
Abstract
Type IV secretion (T4S) systems form the most common and versatile class of secretion systems in bacteria, capable of injecting both proteins and DNAs into host cells. T4S systems are typically composed of 12 components that form 2 major assemblies: the inner membrane complex embedded in the inner membrane and the core complex embedded in both the inner and outer membranes. Here we present the 3.3 Å-resolution cryo-electron microscopy model of the T4S system core complex from Xanthomonas citri, a phytopathogen that utilizes this system to kill bacterial competitors. An extensive mutational investigation was performed to probe the vast network of protein–protein interactions in this 1.13-MDa assembly. This structure expands our knowledge of the molecular details of T4S system organization, assembly and evolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Density map is available at EMDB with accession code EMD-0089. Atomic model is available in Protein Data Bank with accession code 6GYB. NMR data are available at BMRB with accession number 27342. All other data supporting the findings of this study are available from the corresponding authors upon request.
References
Alvarez-Martinez, C. E. & Christie, P. J. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808 (2009).
Odenbreit, S. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287, 1497–1500 (2000).
Isaac, D. T. & Isberg, R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 9, 343–359 (2014).
Kotob, S. I., Hausman, S. Z. & Burns, D. L. Localization of the promoter for the ptl genes of Bordetella pertussis, which encode proteins essential for secretion of pertussis toxin. Infect. Immun. 63, 3227–3230 (1995).
De Jong, M. F. & Tsolis, R. M. Brucellosis and type IV secretion. Future Microbiol. 7, 47–58 (2012).
Dehio, C. & Tsolis, R. M. Type IV effector secretion and subversion of host functions by Bartonella and Brucella species. Curr. Top. Microbiol. Immunol. 413, 269–295 (2017).
Siamer, S. & Dehio, C. New insights into the role of Bartonella effector proteins in pathogenesis. Curr. Opin. Microbiol. 23, 80–85 (2015).
Souza, D. P. et al. Bacterial killing via a type IV secretion system. Nat. Commun. 6, 6453 (2015).
Oliveira, L. C. et al. VirB7 and VirB9 interactions are required for the assembly and antibacterial activity of a type IV secretion system. Structure 24, 1707–1718 (2016).
Chandran, V. & Waksman, G. Structural biology of bacterial type IV secretion systems. Annu. Rev. Biochem. 84, 603–629 (2015).
Low, H. H. et al. Structure of a type IV secretion system. Nature 508, 550–553 (2014).
Costa, T. R. D. et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359 (2015).
Trokter, M., Felisberto-Rodrigues, C., Christie, P. J. & Waksman, G. Recent advances in the structural and molecular biology of type IV secretion systems. Curr. Opin. Struct. Biol. 27, 16–23 (2014).
Fronzes, R. et al. Structure of a type IV secretion system core complex. Science 323, 266–268 (2009).
Chandran, V. et al. Structure of the outer membrane complex of a type IV secretion system. Nature 462, 1011–1015 (2009).
Souza, D. P. et al. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 7, e1002031 (2011).
Rivera-Calzada, A. et al. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 32, 1195–1204 (2013).
Gordon, J. E. et al. Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol. Microbiol. 105, 273–293 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
The PyMOL Molecular Graphics System v.1.8. (Schrödinger, LLC, 2017).
Cavanagh, J., Fairbrother, W. J., Palmer, A. G., Rance, M. & Skelton, N. J. Protein NMR Spectroscopy: Principles and Practice (Academic Press, San Diego, 2007).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Vranken, W. F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct. Funct. Genet. 59, 687–696 (2005).
Marsh, J. A., Singh, V. K., Jia, Z. & Forman-Kay, J. D. Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: implications for fibrillation. Protein Sci. 15, 2795–2804 (2006).
Shen, Y. & Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013).
da Silva, A. C. R. et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463 (2002).
Ke, N., Landgraf, D., Paulsson, J. & Berkmen, M. Visualization of periplasmic and cytoplasmic proteins with a self-labeling protein tag. J. Bacteriol. 198, 1035–1043 (2016).
Sawitzke, J. A. et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J. Mol. Biol. 407, 45–59 (2011).
Vettiger, A., Basler, M. Type VI secretion system substrates are transferred and reused among sister cells. Cell 167, 99–110 (2016).
Clark, D. J. & Maaloe, O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23, 99–112 (1967).
RStudio: Integrated Development for R (RStudio Team, 2016); http://www.rstudio.com/
Wickham, H. ggplot2 (Springer-Verlag, New York, 2009).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2013).
Jiang, C., Brown, P. J. B., Ducret, A. & Brun, Y. V. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506, 489–493 (2014).
Schägger, H. Tricine–SDS–PAGE. Nat. Protoc. 1, 16–22 (2006).
Acknowledgements
This work was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grants 2011/07777-5 and 2017/17303-7 to C.S.F. and Wellcome Trust grant 098302 to G.W. G.G.S. and W.C. were recipients of FAPESP post-doctoral fellowship grants. D.P.S. and L.C.O. received post-doctoral and PhD scholarships, respectively, from the Conselho Nacional de Pesquisa e Desenvolvimento (CNPq). We thank the members of the Farah and Waksman laboratories for fruitful discussions. We thank A. Bruni-Cardoso for fluorescence microscope access. We thank LNNano/CNPEM for the access to the cryo-EM facility, proposals TEM-19470 and TEM-20247. We thank Diamond for access to and support of the Cryo-EM facilities at the UK national Electron Bio-Imaging Centre (eBIC), proposal EM14704, funded by the Wellcome Trust, MRC and BBSRC. We thank N. Lukoyanova for the use of the ISMB Polara microscope.
Author information
Authors and Affiliations
Contributions
G.G.S. cloned, expressed and purified the X. citri T4S system core complex. G.G.S., A.C. and R.V.P. prepared and collected negative-staining EM data. G.G.S., T.R.D.C., A.C. and R.V.P. determined the sample preparation conditions for cryo-EM. T.R.D.C. prepared cryo-EM grids used for data collection (with G.G.S.), collected cryo-EM data, performed the image analysis and carried out the EM reconstructions. G.G.S. built and refined the model. G.G.S., W.C. and D.P.S. carried out the mutagenesis work. W.C. performed and analysed the biological assays and the microscopy analysis. G.G.S. carried out the immunoblotting analysis. L.C.O., D.P.S., C.S.F. and R.K.S. performed the NMR analysis. G.G.S., T.R.D.C., W.C., C.S.F. and G.W. prepared the figures. G.G.S., C.S.F. and G.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–9, Supplementary Tables 1–5, Supplementary Notes, Supplementary Discussion, Supplementary References
Rights and permissions
About this article
Cite this article
Sgro, G.G., Costa, T.R.D., Cenens, W. et al. Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri. Nat Microbiol 3, 1429–1440 (2018). https://doi.org/10.1038/s41564-018-0262-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0262-z