Abstract
Bacteria with increased mutation rates (mutators) are common in chronic infections and are associated with poorer clinical outcomes, especially in the case of Pseudomonas aeruginosa infecting cystic fibrosis (CF) patients. There is, however, considerable between-patient variation in both P. aeruginosa mutator frequency and the composition of co-infecting pathogen communities. We investigated whether community context might affect selection of mutators. Using an in vitro CF model community, we show that P. aeruginosa mutators were favoured in the absence of other species but not in their presence. This was because there were trade-offs between adaptation to the biotic and abiotic environments (for example, loss of quorum sensing and associated toxin production was beneficial in the latter but not the former in our in vitro model community) limiting the evolvability advantage of an elevated mutation rate. Consistent with a role of co-infecting pathogens selecting against P. aeruginosa mutators in vivo, we show that the mutation frequency of P. aeruginosa population was negatively correlated with the frequency and diversity of co-infecting bacteria in CF infections. Our results suggest that co-infecting taxa can select against P. aeruginosa mutators, which may have potentially beneficial clinical consequences.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data used in this study are available on figshare at https://doi.org/10.6084/m9.figshare.13739452. Genome sequencing reads from P. aeruginosa populations from in vivo and in vitro experiments have been deposited under accession no. PRJEB35620. All other data used in this paper are available in the Supplementary Information. Source data are provided with this paper.
References
van Valen, L. A new evolutionary law. Evol. Theory 1, 1–30 (1973).
Stenseth, N. C. & Smith, J. M. Coevolution in ecosystems: Red Queen evolution or stasis? Evolution 38, 870–880 (1984).
de Mazancourt, C., Johnson, E. & Barraclough, T. G. Biodiversity inhibits species’ evolutionary responses to changing environments. Ecol. Lett. 11, 380–388 (2008).
Morran, L. T., Schmidt, O. G., Gelarden, I. A., Parrish, R. C. & Lively, C. M. Running with the Red Queen: host–parasite coevolution selects for biparental sex. Science 333, 216–218 (2011).
Hamilton, W. D., Axelrod, R. & Tanese, R. Sexual reproduction as an adaptation to resist parasites (a review). Proc. Natl Acad. Sci. USA 87, 3566–3573 (1990).
Taddei, F. et al. Role of mutator alleles in adaptive evolution. Nature 387, 700–702 (1997).
Pal, C., Maciá, M. D., Oliver, A., Schachar, I. & Buckling, A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450, 1079–1081 (2007).
terHorst, C. P. et al. Evolution in a community context: trait responses to multiple species interactions. Am. Nat. 191, 368–390 (2018).
Wielgoss, S. et al. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl Acad. Sci. USA 110, 222–227 (2013).
Tenaillon, O., le Nagard, H., Godelle, B. & Taddei, F. Mutators and sex in bacteria: conflict between adaptive strategies. Proc. Natl Acad. Sci. USA 97, 10465–10470 (2000).
Raynes, Y., Wylie, C. S., Sniegowski, P. D. & Weinreich, D. M. Sign of selection on mutation rate modifiers depends on population size. Proc. Natl Acad. Sci. USA 115, 3422–3427 (2018).
LeClerc, J. E., Li, B., Payne, W. L. & Cebula, T. A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274, 1208–1211 (1996).
Matic, I. et al. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277, 1833–1834 (1997).
Denamur, E. et al. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J. Bacteriol. 184, 605–609 (2002).
Oliver, A. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1253 (2000).
Feliziani, S. et al. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS ONE 5, e12669 (2010).
Watson, M. E., Burns, J. L. & Smith, A. L. Hypermutable Haemophilus influenzae with mutations in mutS are found in cystic fibrosis sputum. Microbiology 150, 2947–2958 (2004).
Martina, P. et al. Hypermutation in Burkholderia cepacia complex is mediated by DNA mismatch repair inactivation and is highly prevalent in cystic fibrosis chronic respiratory infection. Int. J. Med. Microbiol. 304, 1182–1191 (2014).
Prunier, A. L. et al. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187, 1709–1716 (2003).
del Campo, R. et al. Population structure, antimicrobial resistance, and mutation frequencies of Streptococcus pneumoniae isolates from cystic fibrosis patients. J. Clin. Microbiol. 43, 2207–2214 (2005).
Jolivet-Gougeon, A. et al. Bacterial hypermutation: clinical implications. J. Med. Microbiol. 60, 563–573 (2011).
Ferroni, A. et al. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis. Pediatr. Pulmonol. 44, 820–825 (2009).
Waine, D. J., Honeybourne, D., Smith, E. G., Whitehouse, J. L. & Dowson, C. G. Association between hypermutator phenotype, clinical variables, mucoid phenotype, and antimicrobial resistance in Pseudomonas aeruginosa. J. Clin. Microbiol. 46, 3491–3493 (2008).
Schaaff, F., Reipert, A. & Bierbaum, G. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46, 3540–3548 (2002).
Maciá, M. D. et al. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 49, 3382–3386 (2005).
Feliziani, S. et al. Coexistence and within-host evolution of diversified lineages of hypermutable Pseudomonas aeruginosa in long-term cystic fibrosis infections. PLoS Genet. 10, e1004651 (2014).
Blázquez, J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 37, 1201–1209 (2003).
Mowat, E. et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183, 1674–1679 (2011).
Harrison, F. Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923 (2007).
O’Brien, S. & Fothergill, J. L. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett. 364, fnx128 (2017).
Alseth, E. O. et al. Bacterial biodiversity drives the evolution of CRISPR-based phage resistance. Nature 574, 549–552 (2019).
Ross-Gillespie, A., Gardner, A., West, S. A. & Griffin, A. S. Frequency dependence and cooperation: theory and a test with bacteria. Am. Nat. 170, 331–342 (2007).
Torres-Barcelo, C., Cabot, G., Oliver, A., Buckling, A. & MacLean, R. C. A trade-off between oxidative stress resistance and DNA repair plays a role in the evolution of elevated mutation rates in bacteria. Proc. R. Soc. B 280, 20130007 (2013).
Chao, L. & Cox, E. C. Competition between high and low mutating strains of Escherichia coli. Evolution 37, 125–134 (1983).
Farrow, J. M. & Pesci, E. C. Distal and proximal promoters co-regulate pqsR expression in Pseudomonas aeruginosa. Mol. Microbiol. 104, 78–91 (2017).
Köhler, T., Ouertatani-Sakouhi, H., Cosson, P. & van Delden, C. QsrO a novel regulator of quorum-sensing and virulence in Pseudomonas aeruginosa. PLoS ONE 9, e87814 (2014).
Tognon, M. et al. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 11, 2233–2243 (2017).
Wright, E. A., Fothergill, J. L., Paterson, S., Brockhurst, M. A. & Winstanley, C. Sub-inhibitory concentrations of some antibiotics can drive diversification of Pseudomonas aeruginosa populations in artificial sputum medium. BMC Microbiol. 13, 170 (2013).
Luján, A. M., Moyano, A. J., Segura, I., Argaraña, C. E. & Smania, A. M. Quorum-sensing-deficient (lasR) mutants emerge at high frequency from a Pseudomonas aeruginosa mutS strain. Microbiology 153, 225–237 (2007).
Heurlier, K. et al. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187, 4875–4883 (2005).
Diggle, S. P., Griffin, A. S., Campbell, G. S. & West, S. A. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414 (2007).
Sandoz, K. M., Mitzimberg, S. M. & Schuster, M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl Acad. Sci. USA 104, 15876–15881 (2007).
Dietrich, L. E. P., Price-Whelan, A., Petersen, A., Whiteley, M. & Newman, D. K. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321 (2006).
Rahme, L. G. et al. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl Acad. Sci. USA 94, 13245–13250 (1997).
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H. & Nomura, N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24 (2013).
Li, C., Wally, H., Miller, S. J. & Lu, C. D. The multifaceted proteins MvaT and MvaU, members of the H-NS family, control arginine metabolism, pyocyanin synthesis, and prophage activation in Pseudomonas aeruginosa PAO1. J. Bacteriol. 191, 6211–6218 (2009).
Rada, B., Gardina, P., Myers, T. G. & Leto, T. L. Reactive oxygen species mediate inflammatory cytokine release and EGFR-dependent mucin secretion in airway epithelial cells exposed to Pseudomonas pyocyanin. Mucosal Immunol. 4, 158–171 (2011).
López-Causapé, C. et al. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci. Rep. 7, 5555 (2017).
Denamur, E. & Matic, I. Evolution of mutation rates in bacteria. Mol. Microbiol. 60, 820–827 (2006).
Giraud, A. et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291, 2606–2608 (2001).
Buckling, A. & Rainey, P. B. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269, 931–936 (2002).
Abdulhussien, Z. R. & Atia, S. S. Antimicrobial effect of pyocyanin extracted from Pseudomonas aeruginosa. Eur. J. Exp. Biol. 6, 6 (2016).
DeBritto, S. et al. Isolation and characterization of nutrient dependent pyocyanin from Pseudomonas aeruginosa and its dye and agrochemical properties. Sci. Rep. 10, 1542 (2020).
Castledine, M. et al. Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. eLife 11, e73679 (2022).
Kirchner, S. et al. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J. Vis. Exp. 64, 3857 (2012).
Davies, E. V., James, C. E., Brockhurst, M. A. & Winstanley, C. Evolutionary diversification of Pseudomonas aeruginosa in an artificial sputum model. BMC Microbiol. 17, 3 (2017).
Choi, K.-H. & Schweizer, H. P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161 (2006).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package version 0.3.30 http://florianhartig.github.io/DHARMa/ (2020).
Lüdecke, D., Ben-Shachar, M. S., Patil, I. & Makowski, D. Extracting, computing and exploring the parameters of statistical models using R. J. Open Source Softw. 5, 2445 (2020).
Lenth, R. V. Least-squares means: the R package lsmeans. J. Stat. Softw. 69, 1–33 (2016).
Wickham, H. ggplot2: Elegant Graphics of Data Analysis (Springer-Verlag, 2016).
Williams, D. et al. Divergent, coexisting Pseudomonas aeruginosa lineages in chronic cystic fibrosis lung infections. Am. J. Respir. Crit. Care Med. https://doi.org/10.1164/rccm.201409-1646OC (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. https://doi.org/10.14806/ej.17.1.200 (2011).
Sarker, M. R. & Cornelis, G. R. An improved version of suicide vector pKNG101 for gene replacement in Gram-negative bacteria. Mol. Microbiol. 23, 410–411 (1997).
Hernandez, R. J. et al. Using the wax moth larva Galleria mellonella infection model to detect emerging bacterial pathogens. PeerJ 6, e6150 (2019).
Therneau, T. M. A Package for Survival Analysis in S. R package version 2.38 http://cran.r-project.org/package=survival (2015).
Lim, Y. W. et al. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J. Cyst. Fibros. 12, 154–164 (2013).
Schmieder, R. & Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 6, e17288 (2011).
Ye, Y., Choi, J.-H. & Tang, H. RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinform. 12, 159 (2011).
Huson, D. H. & Weber, N. Microbial community analysis using MEGAN. Methods Enzymol. 531, 465–485 (2013).
McKenna, A. et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
van der Auwera, G. A. et al. From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11.10.1–11.10.33 (2013).
Acknowledgements
We would like to thank J. P. Pirnay and D. de Vos for providing the bacterial community strains and A. Frias for generating the illustration shown in Extended Data Fig. 1. A.M.L. was supported by FP7 Marie Skłodowska-Curie International Incoming Fellowship (IIF) (no. 331163) and CONICET. E.H. was supported by KRI Future Leaders Fellowship (MR/V022482/1). A.M.S. was funded by ANPCyT (PICT-2016-1545 and PICT-2019-1590). S.M. was supported by Novo Nordisk Foundation Center for Biosustainability (CFB) (NNF10CC1016517). H.K.J. was supported by the Novo Nordisk Foundation as a clinical research stipend (NNF12OC1015920), by Rigshospitalets Rammebevilling 2015–17 (R88-A3537), by Lundbeckfonden (R167-2013-15229), by the Novo Nordisk Foundation (NNF15OC0017444), by RegionH Rammebevilling (R144-A5287) and by Independent Research Fund Denmark/Medical and Health Sciences (FTP-4183-00051). A.B. is supported by NERC (NE/S000771/1 and NE/V012347/1).
Author information
Authors and Affiliations
Contributions
A.M.L. and A.B. conceived and designed the study. A.M.L., L.M.S. and E.O.A. carried out the experiments. S.P., L.M.S., R.L.M. and M.D.S. analysed sequence data. A.M.L. and E.H. analysed experimental data. A.M.L., O.C. and H.K.J. were responsible for the CF sputum sample collection. H.K.J., A.M.S. and S.M. contributed to the design and interpretation of the in vivo analysis. A.M.L. and A.B. wrote the manuscript. All authors provided feedback on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Nic Vega, Jonathan Friedman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
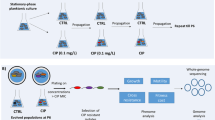
Extended Data Fig. 1
Experimental design of the two complementary in vitro and in vivo approaches carried out in the study.
Extended Data Fig. 2 Parameter estimates and diagnostics plots for the most parsimonious LMM testing the effect of co-culturing on relative mutator fitness (v) through time (Fig. 1a main text).
(a) Table with estimated regression parameters ± 95% confidence intervals (CI), t-statistics and p values. Parameter estimates are on log2-scale. The minimal adequate model was arrived at by sequentially deleting terms and comparing model fits using χ2-tests. The most parsimonious models could be best described by a LMM with co-culturing as sole fixed explanatory variable (treatment: χ2(1) = 5.28, p = 0.02, transfer: χ2(3) = 3.94, p = 0.27 and 2-way interaction: χ2(3) = 1.61, p = 0.66) and random intercepts fitted for each population (estimated value for σPopulation is 1.20). Because we computed a single pairwise contrast (monoculture versus co-culture), we did not adjust p values for multiple testing. (b-c) DHARMa simulation-based residual plots indicate no misspecification in the fitted LMM: (b) QQ-plot showing no significant overall deviations from the expected distribution, (c) Boxplot demonstrating homoscedastic dispersion of residuals across community treatments. Boxplots depict the median, the lower and upper quartiles (the 25th and 75th percentiles), with whiskers extending to 1.5x the interquartile range.
Extended Data Fig. 3 Changes in mutator frequency are driven by indirect selection.
Boxplots depicting temporal changes in relative mutator fitness (𝑣) as a function of community absence and presence. We determined relative mutator fitness (𝑣) by comparing mutator frequencies between consecutive time points, using: 𝑣 = x2(1-x1)/x1(1-x2), where x1 x1 is mutator frequency at transfer (tn) and x2 mutator frequency at transfer (tn+1). Boxes depict the upper and lower quartiles of the treatment-specific raw data with the centre line showing the median and whiskers providing a measure of the 1.5x interquartile range. Dots represent individual replicate populations (n = 12 per community treatment). See Extended Data Fig. 4 for associated model predictions and diagnostics.
Extended Data Fig. 4 Parameter estimates and diagnostics plots of the most parsimonious LMM testing the interactive effect of treatment × transfer (t) on consecutive relative mutator fitness (v) (Extended Data Fig. 3).
(a) Table with estimated regression parameters ± 95% confidence intervals (CI), z- and p values, and pairwise post hoc contrasts. The minimal adequate model was arrived at by sequentially deleting terms and comparing model fits using χ2-tests. The most parsimonious models could be best described by a LMM with transfer as sole fixed explanatory factor (transfer: χ2(3) = 16.75, p < 0.001, treatment: χ2(1) = 1.48, p = 0.22; 2-way interaction: χ2(3) = 0.87, p = 0.83) and random intercepts fitted for each population. Because the estimated value for σPopulation approached zero (0.000049), we fitted the model using the ‘glmmTMB’ R package, using the ‘nlminb’ optimizer and a Gaussian error structure. Note that glmmTMB reports z-statistics (instead of t-statistics) and that parameter estimates are on log2-scale. Pairwise contrasts were corrected for multiple testing using the ‘tukey’ method. (b-c) DHARMa simulation-based residual plots indicate no misspecification in the fitted LMM following model reduction: (b) QQ-plot showing no significant overall deviation from the expected distribution, (c) Boxplot demonstrating homoscedastic dispersion of residuals across time. Boxplots depict the median, the lower and upper quartiles (the 25th and 75th percentiles), with whiskers extending to 1.5x the interquartile range.
Extended Data Fig. 5 Cell densities of P. aeruginosa total population in the absence (monoculture) and in the presence (bacterial community) of the microbial community at the end point of each transfer.
Raw data is shown (n=12). P. aeruginosa population sizes were not affected by the presence of the community (Welch Two Sample two-sided t-test testing the effect of co-culturing on density averaged across time, t = 1.05, df = 22, p = 0.30). Boxplots depict the median, the lower and upper quartiles (the 25th and 75th percentiles), with whiskers extending to 1.5x the interquartile range.
Extended Data Fig. 6 Parameter estimates and diagnostics plots for the fitted LMM testing the presence of a trade-off between biotic versus abiotic adaptation (Fig. 1b in main text).
(a) Table with estimated regression parameters, 95% confidence intervals (CI), t-statistics and p values. We fitted a LMM with relative fitness of evolved PAO1 populations (v) as a response variable, selection × competition environment as fixed explanatory variables and random intercepts fitted for individual populations (estimated value for σPopulation is 1.68). Again, the minimal adequate model was arrived at by sequentially deleting terms and comparing model fits using χ2-tests. The most parsimonious model included the interaction term (selection × competition environment: χ2(1)= 7.07, p = 0.008). To determine whether populations performed better (that is displayed a higher v) when competing with the ancestor under conditions similar to those they had evolved in (that is with or without the community), we carried out pairwise contrasts between competition treatments within each of the two selection environments. Hence, p values were not adjusted for multiple testing and estimated using the kenward-roger degrees-of-freedom method. Parameter estimates are on log2-scale. (b) We confirmed the robustness of our model predictions using a Bayesian framework (warmup = 1000, post warmup = 4000, chains = 4, thin = 1 and weakly informed priors), which gave very similar parameter estimates. (c-e) DHARMa simulation-based residual plots indicate no misspecification in the fitted LMM (see Fig. 1b for raw data). (c) QQ-plot showing no significant overall deviations from the expected distribution, and (d-e) Boxplots demonstrating homoscedastic dispersion of residuals across selection regimes and competition environments, respectively. Boxplots depict the median, the lower and upper quartiles (the 25th and 75th percentiles), with whiskers extending to 1.5x the interquartile range.
Extended Data Fig. 7 Parameter estimates and diagnostics plots for the fitted GLM testing the effect of community presence on mutant fitness (v) (strains ∆mvaU versus ∆pqsR), relative to the ancestral strain (Fig. 2b in main text).
(a) Table with estimated regression parameters ± 95% confidence intervals (CI), t-statistics and p values. The minimal adequate model was arrived at by sequentially deleting terms and comparing model fits using F-tests. The most parsimonious model included the interaction term (mutant strain × community context: F1, 20 = 13.21, p = 0.002). We carried out pairwise contrasts between strains within each of the two competition environments; hence, p values were not adjusted for multiple testing (see method section in the main text). Parameter estimates are on log2-scale. (b-d) DHARMa simulation-based residual plots indicate no misspecification in the fitted GLM. (b) QQ-plot showing no significant overall deviations from the expected distribution, and (c-d) Boxplots demonstrating homoscedastic dispersion of residuals across community treatments and strains, respectively. Boxplots depict the median, the lower and upper quartiles (the 25th and 75th percentiles), with whiskers extending to 1.5x the interquartile range.
Extended Data Fig. 8 Relation between P. aeruginosa average mutation frequency and the number of CF resistome genes fixed (>0.5) in vivo.
Each point represents data obtained for each P. aeruginosa population obtained from the twenty-four CF patients (two-sided Pearson’s t = 0.98t, df = 22, p-value = 0.3344).
Supplementary information
Source data
Source Data Fig. 1
Statistical source data for Fig. 1.
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Figs. 3.
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5.
Source Data Extended Data Fig. 8
Statistical source data for Extended Data Fig. 8.
Rights and permissions
About this article
Cite this article
Luján, A.M., Paterson, S., Hesse, E. et al. Polymicrobial infections can select against Pseudomonas aeruginosa mutators because of quorum-sensing trade-offs. Nat Ecol Evol 6, 979–988 (2022). https://doi.org/10.1038/s41559-022-01768-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-022-01768-1
This article is cited by
-
Recent advances in therapeutic targets identification and development of treatment strategies towards Pseudomonas aeruginosa infections
BMC Microbiology (2023)
-
Impact of transient acquired hypermutability on the inter- and intra-species competitiveness of Pseudomonas aeruginosa
The ISME Journal (2023)