Abstract
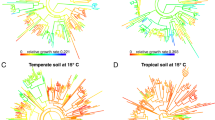
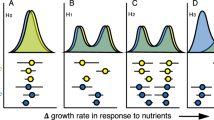
Organisms influence ecosystems, from element cycling to disturbance regimes, to trophic interactions and to energy partitioning. Microorganisms are part of this influence, and understanding their ecology in nature requires studying the traits of these organisms quantitatively in their natural habitats—a challenging task, but one which new approaches now make possible. Here, we show that growth rate and carbon assimilation rate of soil microorganisms are influenced more by evolutionary history than by climate, even across a broad climatic gradient spanning major temperate life zones, from mixed conifer forest to high-desert grassland. Most of the explained variation (~50% to ~90%) in growth rate and carbon assimilation rate was attributable to differences among taxonomic groups, indicating a strong influence of evolutionary history, and taxonomic groupings were more predictive for organisms responding to resource addition. With added carbon and nitrogen substrates, differences among taxonomic groups explained approximately eightfold more variance in growth rate than did differences in ecosystem type. Taxon-specific growth and carbon assimilation rates were highly intercorrelated across the four ecosystems, constrained by the taxonomic identity of the organisms, such that plasticity driven by environment was limited across ecosystems varying in temperature, precipitation and dominant vegetation. Taken together, our results suggest that, similar to multicellular life, the traits of prokaryotes in their natural habitats are constrained by evolutionary history to a greater degree than environmental variation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Victor O. Leshyk, Center for Ecosystem Science and Society, Northern Arizona University




Similar content being viewed by others
Data availability
The data supporting this manuscript are available in the NCBI short-read archive under accession no. PRJNA521534.
References
Sitch, S. et al. Evaluation of ecosystem dynamics, plant geography and terrestrial carbon cycling in the LPJ dynamic global vegetation model. Glob. Change Biol. 9, 161–185 (2003).
Verheijen, L. M. et al. Inclusion of ecologically based trait variation in plant functional types reduces the projected land carbon sink in an earth system model. Glob. Change Biol. 21, 3074–3086 (2015).
Krause, S. et al. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5, 251 (2014).
Hungate, B. A. et al. Quantitative microbial ecology through stable isotope probing. Appl. Environ. Microbiol. 81, 7570–7581 (2015).
Tripathi, A. et al. Are microbiome studies ready for hypothesis-driven research? Curr. Opin. Microbiol. 44, 61–69 (2018).
Martiny, A. C., Treseder, K. & Pusch, G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7, 830–838 (2013).
Ho, A., Di Lonardo, D. P. & Bodelier, P. L. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93, fix006 (2017).
Green, J. L., Bohannan, B. J. & Whitaker, R. J. Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043 (2008).
Philippot, L. et al. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8, 523 (2010).
McGill, B. J. Exploring predictions of abundance from body mass using hierarchical comparative approaches. Am. Nat. 172, 88–101 (2008).
Neyret, M. et al. Examining variation in the leaf mass per area of dominant species across two contrasting tropical gradients in light of community assembly. Ecol. Evol. 6, 5674–5689 (2016).
Messier, J., McGill, B. J. & Lechowicz, M. J. How do traits vary across ecological scales? A case for trait‐based ecology. Ecol. Lett. 13, 838–848 (2010).
Cornwell, W. K. & Ackerly, D. D. Community assembly and shifts in plant trait distributions across an environmental gradient in coastal California. Ecol. Monogr. 79, 109–126 (2009).
McGill, B. J. et al. Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185 (2006).
Martiny, J. B. et al. Microbiomes in light of traits: a phylogenetic perspective. Science 350, aac9323 (2015).
Gogarten, J. P. & Townsend, J. P. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3, 679 (2005).
Popa, O., Landan, G. & Dagan, T. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 11, 543 (2017).
Lenski, R. E. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 11, 2181 (2017).
Blankinship, J. C. et al. Response of terrestrial CH4 uptake to interactive changes in precipitation and temperature along a climatic gradient. Ecosystem 13, 1157–1170 (2010).
Liu, X. J. A. et al. Labile carbon input determines the direction and magnitude of the priming effect. Appl. Soil Ecol. 109, 7–13 (2017).
Kuzyakov, Y. & Blagodatskaya, E. Microbial hotspots and hot moments in soil: concept & review. Soil Biol. Biochem. 83, 184–199 (2015).
Jones, D. L., Nguyen, C. & Finlay, R. D. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321, 5–33 (2009).
Austin, A. T. Water pulses and biogeochemical cycles in arid and semiarid ecosystems. Oecologia 141, 221–235 (2004).
Leff, J. W. et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl Acad. Sci. USA 112, 10967–10972 (2015).
Albert, C. H. et al. A multi‐trait approach reveals the structure and the relative importance of intra‐vs. interspecific variability in plant traits. Funct. Ecol. 24, 1192–1201 (2010).
Jiang, Y. et al. Interspecific and intraspecific variation in functional traits of subtropical evergreen and deciduous broadleaved mixed forests in Karst topography, Guilin, Southwest China. Trop. Conserv. Sci. 9 9, https://doi.org/10.1177/1940082916680211 (2016).
Yarza, P. et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635 (2014).
Morrissey, E. M. et al. Phylogenetic organization of bacterial activity. ISME J. 10, 2016 (2016).
Goberna, M. & Verdú, M. Predicting microbial traits with phylogenies. ISME J. 10, 959 (2016).
Vilà‐Cabrera, A., Martínez‐Vilalta, J. & Retana, J. Functional trait variation along environmental gradients in temperate and Mediterranean trees. Glob. Ecol. Biogeogr. 24, 1377–1389 (2015).
Allison, S. D. & Martiny, J. B. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl Acad. Sci. USA 105, 11512–11519 (2008).
Morrissey, E. M. et al. Taxonomic patterns in the nitrogen assimilation of soil prokaryotes. Environ. Microbiol. 20, 1112–1119 (2018).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621 (2012).
Berry, D. et al. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 77, 7846–7849 (2011).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335 (2010).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Caporaso, J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010).
Wang, Q. et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Bokulich, N. A. et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57 (2013).
Morrissey, E. M. et al. (2017). Bacterial carbon use plasticity, phylogenetic diversity and the priming of soil organic matter. ISME J. 11, 1890 (2017).
Acknowledgements
This research was supported by grants from the Department of Energy’s Biological Systems Science Division (No. DE-SC0016207), Program in Genomic Science and the National Science Foundation (Nos. DEB-1645596 and DEB-1241094). Work at Lawrence Livermore National Laboratory (LLNL) was funded by the Department of Energy through the Genome Sciences Program under contract Nos. SCW1424 and SCW1590, and performed under the auspices of LLNL under Contract No. DE-AC52-07NA27344.
Author information
Authors and Affiliations
Contributions
E.M.M., E.S. and B.A.H. developed the core ideas explored in this manuscript. R.L.M., M.H. and X.-J.A.L. carried out the soil manipulation and participated in sample analysis. R.L.M. and M.H. performed the sequencing and initial bioinformatics analysis. E.M.M., B.J.K. and K.A. performed the data analysis. K.H., J.P-R., S.J.B., B.A.H. and P.D. provided guidance on data analysis and interpretation. E.M.M. drafted the manuscript and all authors contributed equally to writing, editing and revision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Supplementary Figs. 1–3
Rights and permissions
About this article
Cite this article
Morrissey, E.M., Mau, R.L., Hayer, M. et al. Evolutionary history constrains microbial traits across environmental variation. Nat Ecol Evol 3, 1064–1069 (2019). https://doi.org/10.1038/s41559-019-0918-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0918-y
This article is cited by
-
Experimental warming accelerates positive soil priming in a temperate grassland ecosystem
Nature Communications (2024)
-
Role in aromatic metabolites biodegradation and adverse implication of denitrifying microbiota in kitchen waste composting
Environmental Microbiome (2023)
-
The predictive power of phylogeny on growth rates in soil bacterial communities
ISME Communications (2023)
-
Rapid growth rate responses of terrestrial bacteria to field warming on the Antarctic Peninsula
The ISME Journal (2023)
-
Functional comparison of metabolic networks across species
Nature Communications (2023)