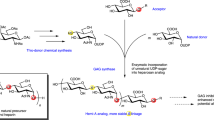
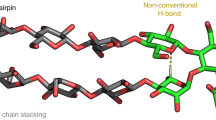
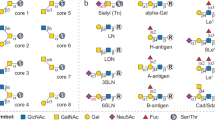
Abstract
Glycosaminoglycans (GAGs) are abundant, ubiquitous carbohydrates in biology, yet their structural complexity has limited an understanding of their biological roles and structure–function relationships. Synthetic access to large collections of well defined, structurally diverse GAG oligosaccharides would provide critical insights into this important class of biomolecules and represent a major advance in glycoscience. Here we report a new platform for synthesizing large heparan sulfate (HS) oligosaccharide libraries displaying comprehensive arrays of sulfation patterns. Library synthesis is made possible by improving the overall synthetic efficiency through universal building blocks derived from natural heparin and a traceless fluorous tagging method for rapid purification with minimal manual manipulation. Using this approach, we generated a complete library of 64 HS oligosaccharides displaying all possible 2-O-, 6-O- and N-sulfation sequences in the tetrasaccharide GlcN–IdoA–GlcN–IdoA. These diverse structures provide an unprecedented view into the sulfation code of GAGs and identify sequences for modulating the activities of important growth factors and chemokines.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data associated with this report are contained within the manuscript or the Supplementary Information. Source data are provided with this Paper.
Code availability
All code associated with this report is publicly available at https://doi.org/10.5281/zenodo.7787616.
References
Zhang, P. et al. Heparan sulfate organizes neuronal synapses through neurexin partnerships. Cell 174, 1450–1464 (2018).
Taylor, K. R. & Gallo, R. L. Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 20, 9–22 (2006).
Parish, C. R. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 6, 633–643 (2006).
Bulow, H. E. & Hobert, O. The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375–407 (2006).
Kusche-Gullberg, M. & Kjellen, L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 13, 605–611 (2003).
Vallet, S. D., Berthollier, C. & Ricard-Blum, S. The glycosaminoglycan interactome 2.0. Am. J. Physiol. Cell Physiol. 322, C1271–C1278 (2022).
Chanalaris, A. et al. Heparan sulfate proteoglycan synthesis is dysregulated in human osteoarthritic cartilage. Am. J. Pathol. 189, 632–647 (2019).
Gordts, P. & Esko, J. D. The heparan sulfate proteoglycan grip on hyperlipidemia and atherosclerosis. Matrix Biol. 71–72, 262–282 (2018).
Soares da Costa, D., Reis, R. L. & Pashkuleva, I. Sulfation of glycosaminoglycans and its implications in human health and disorders. Annu. Rev. Biomed. Eng. 19, 1–26 (2017).
Mah, D. et al. The sulfation code of tauopathies: heparan sulfate proteoglycans in the prion like spread of tau pathology. Front. Mol. Biosci. 8, 671458 (2021).
Mende, M. et al. Chemical synthesis of glycosaminoglycans. Chem. Rev. 116, 8193–8255 (2016).
Zong, C. et al. Integrated approach to identify heparan sulfate ligand requirements of Robo1. J. Am. Chem. Soc. 138, 13059–13067 (2016).
Zulueta, M. M. et al. α-Glycosylation by d-glucosamine-derived donors: synthesis of heparosan and heparin analogues that interact with mycobacterial heparin-binding hemagglutinin. J. Am. Chem. Soc. 134, 8988–8995 (2012).
Noti, C., de Paz, J. L., Polito, L. & Seeberger, P. H. Preparation and use of microarrays containing synthetic heparin oligosaccharides for the rapid analysis of heparin-protein interactions. Chem. Eur. J. 12, 8664–8686 (2006).
Ramadan, S. et al. Automated solid phase assisted synthesis of a heparan sulfate disaccharide library. Org. Chem. Front. 9, 2910–2920 (2022).
Hu, Y. P. et al. Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction. J. Am. Chem. Soc. 134, 20722–20727 (2012).
Jain, P. et al. Discovery of rare sulfated N-unsubstituted glucosamine based heparan sulfate analogs selectively activating chemokines. Chem. Sci. 12, 3674–3681 (2021).
Pawar, N. J. et al. Expedient synthesis of core disaccharide building blocks from natural polysaccharides for heparan sulfate oligosaccharide assembly. Angew. Chem. Int. Ed. 58, 18577–18583 (2019).
Wu, B. et al. Facile chemoenzymatic synthesis of biotinylated heparosan hexasaccharide. Org. Biomol. Chem. 13, 5098–5101 (2015).
Hansen, S. U. et al. Tetrasaccharide iteration synthesis of a heparin-like dodecasaccharide and radiolabelling for in vivo tissue distribution studies. Nat. Commun. 4, 2016 (2013).
Chen, Y. et al. Tailored design and synthesis of heparan sulfate oligosaccharide analogues using sequential one-pot multienzyme systems. Angew. Chem. Int. Ed. 52, 11852–11856 (2013).
Xu, Y. et al. Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science 334, 498–501 (2011).
Hu, Y. P. et al. Synthesis of 3-O-sulfonated heparan sulfate octasaccharides that inhibit the herpes simplex virus type 1 host–cell interaction. Nat. Chem. 3, 557–563 (2011).
Wang, Z. et al. Preactivation-based, one-pot combinatorial synthesis of heparin-like hexasaccharides for the analysis of heparin-protein interactions. Chem. Eur. J. 16, 8365–8375 (2010).
Chen, C. & Yu, B. Efficient synthesis of Idraparinux, the anticoagulant pentasaccharide. Bioorg. Med. Chem. Lett. 19, 3875–3879 (2009).
Dey, S. & Wong, C. H. Programmable one-pot synthesis of heparin pentasaccharides enabling access to regiodefined sulfate derivatives. Chem. Sci. 9, 6685–6691 (2018).
Zong, C. et al. Heparan sulfate microarray reveals that heparan sulfate-protein binding exhibits different ligand requirements. J. Am. Chem. Soc. 139, 9534–9543 (2017).
Hansen, S. U. et al. Synthesis of l-iduronic acid derivatives via [3.2.1] and [2.2.2] l-iduronic lactones from bulk glucose-derived cyanohydrin hydrolysis: a reversible conformationally switched superdisarmed/rearmed lactone route to heparin disaccharides. J. Org. Chem. 80, 3777–3789 (2015).
Codee, J. D. et al. A modular strategy toward the synthesis of heparin-like oligosaccharides using monomeric building blocks in a sequential glycosylation strategy. J. Am. Chem. Soc. 127, 3767–3773 (2005).
Munoz-Garcia, J. C. et al. Effect of the substituents of the neighboring ring in the conformational equilibrium of iduronate in heparin-like trisaccharides. Chem. Eur. J. 18, 16319–16331 (2012).
Panza, M., Pistorio, S. G., Stine, K. J. & Demchenko, A. V. Automated chemical oligosaccharide synthesis: novel approach to traditional challenges. Chem. Rev. 118, 8105–8150 (2018).
Hahm, H. S. et al. Automated glycan assembly of oligo-N-acetyllactosamine and keratan sulfate probes to study virus-glycan interactions. Chem 2, 114–124 (2017).
Eller, S., Collot, M., Yin, J., Hahm, H. S. & Seeberger, P. H. Automated solid-phase synthesis of chondroitin sulfate glycosaminoglycans. Angew. Chem. Int. Ed. 52, 5858–5861 (2013).
Budhadev, D. et al. Using automated glycan assembly (AGA) for the practical synthesis of heparan sulfate oligosaccharide precursors. Org. Biomol. Chem. 17, 1817–1821 (2019).
Walvoort, M. T. et al. Automated solid-phase synthesis of hyaluronan oligosaccharides. Org. Lett. 14, 3776–3779 (2012).
Raman, R., Venkataraman, G., Ernst, S., Sasisekharan, V. & Sasisekharan, R. Structural specificity of heparin binding in the fibroblast growth factor family of proteins. Proc. Natl Acad. Sci. USA 100, 2357–2362 (2003).
Xu, D. & Esko, J. D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83, 129–157 (2014).
Guglier, S. et al. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochem. 47, 13862–13869 (2008).
Shaw, J. P. et al. The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure 12, 2081–2093 (2004).
Studer, A. et al. Fluorous synthesis: a fluorous-phase strategy for improving separation efficiency in organic synthesis. Science 275, 823–826 (1997).
Kern, M. K. & Pohl, N. L. B. Automated solution-phase synthesis of S-glycosides for the production of oligomannopyranoside derivatives. Org. Lett. 22, 4156–4159 (2020).
Bhaduri, S. & Pohl, N. L. Fluorous-tag assisted syntheses of sulfated keratan sulfate oligosaccharide fragments. Org. Lett. 18, 1414–1417 (2016).
Cai, C. et al. Fluorous-assisted chemoenzymatic synthesis of heparan sulfate oligosaccharides. Org. Lett. 16, 2240–2243 (2014).
Zong, C., Venot, A., Dhamale, O. & Boons, G. J. Fluorous supported modular synthesis of heparan sulfate oligosaccharides. Org. Lett. 15, 342–345 (2013).
Jaipuri, F. A. & Pohl, N. L. Toward solution-phase automated iterative synthesis: fluorous-tag assisted solution-phase synthesis of linear and branched mannose oligomers. Org. Biomol. Chem. 6, 2686–2691 (2008).
Bock, K. & Pedersen, C. A study of 13CH coupling constants in hexopyranoses. J. Chem. Soc. Perkin Trans. 2, 293–297 (1974).
Ishihara, M., Kariya, Y., Kikuchi, H., Minamisawa, T. & Yoshida, K. Importance of 2-O-sulfate groups of uronate residues in heparin for activation of FGF-1 and FGF-2. J. Biochem. 121, 345–349 (1997).
Guimond, S., Maccarana, M., Olwin, B. B., Lindahl, U. & Rapraeger, A. C. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2 and FGF-4. J. Biol. Chem. 268, 23906–23914 (1993).
Ashikari-Hada, S. et al. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem. 279, 12346–12354 (2004).
Beenken, A. & Mohammadi, M. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug. Discov. 8, 235–253 (2009).
Schneider, T. D. & Stephens, R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Faham, S., Hileman, R. E., Fromm, J. R., Linhardt, R. J. & Rees, D. C. Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120 (1996).
Faham, S., Linhardt, R. J. & Rees, D. C. Diversity does make a difference: fibroblast growth factor-heparin interactions. Curr. Opin. Struct. Biol. 8, 578–586 (1998).
Ishihara, M. Structural requirements in heparin for binding and activation of FGF-1 and FGF-4 are different from that for FGF-2. Glycobiology 4, 817–824 (1994).
Vora, S. M., Lieberman, J. & Wu, H. Inflammasome activation at the crux of severe COVID-19. Nat. Rev. Immunol. 21, 694–703 (2021).
Moore, B. B. & Kunkel, S. L. Attracting attention: discovery of IL-8/CXCL8 and the birth of the chemokine field. J. Immunol. 202, 3–4 (2019).
de Paz, J. L. et al. Profiling heparin-chemokine interactions using synthetic tools. ACS Chem. Biol. 2, 735–744 (2007).
Spillmann, D., Witt, D. & Lindahl, U. Defining the interleukin-8-binding domain of heparan sulfate. J. Biol. Chem. 273, 15487–15493 (1998).
Rawat, M., Gama, C. I., Matson, J. B. & Hsieh-Wilson, L. C. Neuroactive chondroitin sulfate glycomimetics. J. Am. Chem. Soc. 130, 2959–2961 (2008).
Sheng, G. J., Oh, Y. I., Chang, S. K. & Hsieh-Wilson, L. C. Tunable heparan sulfate mimetics for modulating chemokine activity. J. Am. Chem. Soc. 135, 10898–10901 (2013).
Oh, Y. I., Sheng, G. J., Chang, S. K. & Hsieh-Wilson, L. C. Tailored glycopolymers as anticoagulant heparin mimetics. Angew. Chem. Int. Ed. 52, 11796–11799 (2013).
Naticchia, M. R., Laubach, L. K., Honigfort, D. J., Purcell, S. C. & Godula, K. Spatially controlled glycocalyx engineering for growth factor patterning in embryoid bodies. Biomater. Sci. 9, 1652–1659 (2021).
Loka, R. S. et al. Heparan sulfate mimicking glycopolymer prevents pancreatic β cell destruction and suppresses inflammatory cytokine expression in islets under the challenge of upregulated heparanase. ACS Chem. Biol. 17, 1387–1400 (2022).
Acknowledgements
Financial support was provided by the National Institutes of Health (NIH U01 GM116262 to L.C.H.-W. and X.H., R44 GM134738 to V. Pagadala, L.C.H.-W. and X.H., U01 GM116248 to N.L.B.P., and T32 GM109825 and T32 GM131994 to M.K.K.) and the National Science Foundation (DGE-1745301 to A.W.S.). We thank M. Shahgholi in the CCE Division Mass Spectrometry Facility and D. Vander Velde in the CCE Division NMR Facility at Caltech for technical support.
Author information
Authors and Affiliations
Contributions
L.W., A.W.S., B.-S.H., N.P., X.H., J.L., N.L.B.P. and L.C.H.-W. designed the research. L.W., A.W.S., B.-S.H., M.K.K. and G.S. performed the research. L.W., A.W.S., B.-S.H. and L.C.H.-W. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
J.L. is a founder and chief scientific officer of Glycan Therapeutics. G.S. is an employee of Glycan Therapeutics and has stock options from the company. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Hongzhi Cao, Chi-Huey Wong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1 and 2, synthetic procedures and characterization, and NMR spectra.
Source data
Source Data Fig. 4
Microarray data provided as fluorescence signal for FGF2.
Source Data Fig. 6
Microarray data provided as fluorescence signal for FGF4.
Source Data Fig. 6
Microarray data provided as fluorescence signal for CXCL8.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, L., Sorum, A.W., Huang, BS. et al. Efficient platform for synthesizing comprehensive heparan sulfate oligosaccharide libraries for decoding glycosaminoglycan–protein interactions. Nat. Chem. 15, 1108–1117 (2023). https://doi.org/10.1038/s41557-023-01248-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-023-01248-4
This article is cited by
-
Dimethyl sulfate and diisopropyl sulfate as practical and versatile O-sulfation reagents
Nature Communications (2024)