Abstract
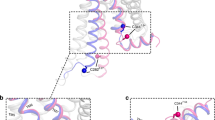
Recent high-pressure NMR results indicate that the preactive conformation of the β1-adrenergic receptor (β1AR) harbours completely empty cavities of ~100 Å3 volume, which disappear in the active conformation of the receptor. Here we have localized these cavities using X-ray crystallography of xenon-derivatized β1AR crystals. One of the cavities is in direct contact with the cholesterol-binding pocket. Solution NMR shows that addition of the cholesterol analogue cholesteryl hemisuccinate impedes the formation of the active conformation of detergent-solubilized β1AR by blocking conserved G protein-coupled receptor microswitches, concomitant with an affinity reduction of both isoprenaline and G protein-mimicking nanobody Nb80 for β1AR detected by isothermal titration calorimetry. This wedge-like action explains the function of cholesterol as a negative allosteric modulator of β1AR. A detailed understanding of G protein-coupled receptor regulation by cholesterol by filling of a dry void and the easy scouting for such voids by xenon may provide new routes for the development of allosteric drugs.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
NMR spectra and ITC raw data, as well as structure factors, phases and density maps derived from the anomalous scattering data of the four xenon-derivatized isoprenaline·β1AR crystals and the β1AR structure derived from the first crystal have been deposited in the Zenodo repository under https://doi.org/10.5281/zenodo.4926013. Source data are provided with this paper.
References
Hubbard, S. J., Gross, K.-H. & Argos, P. Intramolecular cavities in globular proteins. Protein Eng. Des. Sel. 7, 613–626 (1994).
Williams, M. A., Goodfellow, J. M. & Thornton, J. M. Buried waters and internal cavities in monomeric proteins. Protein Sci. 3, 1224–1235 (1994).
Otting, G., Liepinsh, E. & Wuthrich, K. Protein hydration in aqueous solution. Science 254, 974–980 (1991).
Desvaux, H. et al. Dynamics of xenon binding inside the hydrophobic cavity of pseudo-wild-type bacteriophage T4 lysozyme explored through xenon-based NMR spectroscopy. J. Am. Chem. Soc. 127, 11676–11683 (2005).
Krimmer, S. G., Cramer, J., Schiebel, J., Heine, A. & Klebe, G. How nothing boosts affinity: hydrophobic ligand binding to the virtually vacated S1′ pocket of thermolysin. J. Am. Chem. Soc. 139, 10419–10431 (2017).
Qvist, J., Davidovic, M., Hamelberg, D. & Halle, B. A dry ligand-binding cavity in a solvated protein. Proc. Natl Acad. Sci. USA 105, 6296–6301 (2008).
Otting, G., Liepinsh, E., Halle, B. & Frey, U. NMR identification of hydrophobic cavities with low water occupancies in protein structures using small gas molecules. Nat. Struct. Mol. Biol. 4, 396–404 (1997).
Abiko, L. A., Grahl, A. & Grzesiek, S. High pressure shifts the β1-adrenergic receptor to the active conformation in the absence of G protein. J. Am. Chem. Soc. 141, 16663–16670 (2019).
Alexander, S. P. et al. The concise guide to pharmacology 2017/18: G protein-coupled receptors. Br. J. Pharmacol. 174, S17–S129 (2017).
Grahl, A., Abiko, L. A., Isogai, S., Sharpe, T. & Grzesiek, S. A high-resolution description of β1-adrenergic receptor functional dynamics and allosteric coupling from backbone NMR. Nat. Commun. 11, 2216 (2020).
Isogai, S. et al. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016).
Kofuku, Y. et al. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 3, 1045 (2012).
Latorraca, N. R., Venkatakrishnan, A. J. & Dror, R. O. GPCR dynamics: structures in motion. Chem. Rev. 117, 139–155 (2017).
Liu, J. J., Horst, R., Katritch, V., Stevens, R. C. & Wüthrich, K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 (2012).
Manglik, A. & Kobilka, B. The role of protein dynamics in GPCR function: insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 27, 136–143 (2014).
Okude, J. et al. Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the μ-opioid receptor. Angew. Chem. Int. Ed. 54, 15771–15776 (2015).
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013).
Ye, L., Van Eps, N., Zimmer, M., Ernst, O. P. & Prosser, R.S. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533, 265–268 (2016).
Rasmussen, S. G. F. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Ballesteros, J. A. & Weinstein, H. in Methods in Neurosciences Vol. 25 (ed. Sealfon, S. C.) 366–428 (Academic Press, 1995).
Trzaskowski, B. et al. Action of molecular switches in GPCRs - theoretical and experimental studies. Curr. Med. Chem. 19, 1090–1109 (2012).
Miller, J. L. & Tate, C. G. Engineering an ultra-thermostable β1-adrenoceptor. J. Mol. Biol. 413, 628–638 (2011).
Miller-Gallacher, J. L. et al. The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS One 9, e92727 (2014).
Heydenreich, F. M., Vuckovic, Z., Matkovic, M. & Veprintsev, D. B. Stabilization of G protein-coupled receptors by point mutations. Front. Pharmacol. 6, 82 (2015).
Warne, T. et al. Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 (2008).
Goncalves, J. A. et al. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl Acad. Sci. USA 107, 19861–19866 (2010).
Manglik, A. & Kruse, A. C. Structural basis for G protein-coupled receptor activation. Biochemistry 56, 5628–5634 (2017).
Oates, J. & Watts, A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol. 21, 802–807 (2011).
Dawaliby, R. et al. Allosteric regulation of G protein–coupled receptor activity by phospholipids. Nat. Chem. Biol. 12, 35–39 (2016).
Salas-Estrada, L. A., Leioatts, N., Romo, T. D. & Grossfield, A. Lipids alter rhodopsin function via ligand-like and solvent-like interactions. Biophys. J. 114, 355–367 (2018).
Song, W., Yen, H.-Y., Robinson, C. V. & Sansom, M. S. P. State-dependent lipid interactions with the A2a receptor revealed by MD simulations using in vivo-mimetic membranes. Structure 27, 392–403.e3 (2019).
Duncan, A. L., Song, W. & Sansom, M. S. P. Lipid-dependent regulation of ion channels and G protein–coupled receptors: insights from structures and simulations. Annu. Rev. Pharmacol. Toxicol. 60, 31–50 (2020).
Pucadyil, T. J. & Chattopadhyay, A. Role of cholesterol in the function and organization of G-protein coupled receptors. Prog. Lipid Res. 45, 295–333 (2006).
Paila, Y. D. & Chattopadhyay, A. in Cholesterol Binding and Cholesterol Transport Proteins: Structure and Function in Health and Disease (ed. Harris, J. R.) 439–466 (Springer Netherlands, 2010); https://doi.org/10.1007/978-90-481-8622-8_16
Escribá, P. V. et al. Membrane lipid therapy: modulation of the cell membrane composition and structure as a molecular base for drug discovery and new disease treatment. Prog. Lipid Res. 59, 38–53 (2015).
Gimpl, G. Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids 199, 61–73 (2016).
Jafurulla, Md., Aditya Kumar, G., Rao, B. D. & Chattopadhyay, A. A in Cholesterol Modulation of Protein Function: Sterol Specificity and Indirect Mechanisms (eds. Rosenhouse-Dantsker, A. & Bukiya, A. N.) 21–52 (Springer International Publishing, 2019); https://doi.org/10.1007/978-3-030-04278-3_2
Kiriakidi, S. et al. in Direct Mechanisms in Cholesterol Modulation of Protein Function (eds. Rosenhouse-Dantsker, A. & Bukiya, A. N.) 89–103 (Springer International Publishing, 2019); https://doi.org/10.1007/978-3-030-14265-0_5
Casares, D., Escribá, P. V. & Rosselló, C. A. Membrane lipid composition: effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 20, doi:10.3390/ijms20092167, (2019).
Jakubík, J. & El-Fakahany, E. E. Allosteric modulation of GPCRs of class A by cholesterol. Int. J. Mol. Sci. 22, 1953 (2021).
Albert, A. D., Boesze-Battaglia, K., Paw, Z., Watts, A. & Epand, R. M. Effect of cholesterol on rhodopsin stability in disk membranes. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol. 1297, 77–82 (1996).
Gimpl, G. & Fahrenholz, F. Cholesterol as stabilizer of the oxytocin receptor. Biochim. Biophys. Acta - Biomembr. 1564, 384–392 (2002).
Yao, Z. & Kobilka, B. Using synthetic lipids to stabilize purified β2 adrenoceptor in detergent micelles. Anal. Biochem. 343, 344–346 (2005).
Jaakola, V.-P. et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 (2008).
Saxena, R. & Chattopadhyay, A. Membrane cholesterol stabilizes the human serotonin1A receptor. Biochim. Biophys. Acta - Biomembr. 1818, 2936–2942 (2012).
Zocher, M., Zhang, C., Rasmussen, S. G. F., Kobilka, B. K. & Müller, D. J. Cholesterol increases kinetic, energetic, and mechanical stability of the human β2-adrenergic receptor. Proc. Natl Acad. Sci. USA 109, E3463–E3472 (2012).
Abiko, L. A., Rogowski, M., Gautier, A., Schertler, G. & Grzesiek, S. Efficient production of a functional G protein-coupled receptor in E. coli for structural studies. J. Biomol. NMR 75, 25–38 (2021).
Bari, M., Paradisi, A., Pasquariello, N. & Maccarrone, M. Cholesterol-dependent modulation of type 1 cannabinoid receptors in nerve cells. J. Neurosci. Res. 81, 275–283 (2005).
Harikumar, K. G. et al. Differential effects of modification of membrane cholesterol and sphingolipids on the conformation, function, and trafficking of the G protein-coupled cholecystokinin receptor. J. Biol. Chem. 280, 2176–2185 (2005).
Muth, S., Fries, A. & Gimpl, G. Cholesterol-induced conformational changes in the oxytocin receptor. Biochem. J. 437, 541–553 (2011).
Qiu, Y., Wang, Y., Law, P.-Y., Chen, H.-Z. & Loh, H. H. Cholesterol regulates μ-opioid receptor-induced β-arrestin 2 translocation to membrane lipid rafts. Mol. Pharmacol. 80, 210–218 (2011).
Casiraghi, M. et al. Functional modulation of a G protein-coupled receptor conformational landscape in a lipid bilayer. J. Am. Chem. Soc. 138, 11170–11175 (2016).
Manna, M. et al. Mechanism of allosteric regulation of β2-adrenergic receptor by cholesterol. eLife 5, e18432 (2016).
Hanson, M. A. et al. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905 (2008).
Liu, W. et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236 (2012).
Wacker, D. et al. Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 (2013).
Manglik, A. et al. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326 (2012).
Zhang, K. et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509, 115–118 (2014).
Wu, H. et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014).
Burg, J. S. et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein–coupled receptor. Science 347, 1113–1117 (2015).
Taghon, G. J., Rowe, J. B., Kapolka, N. J. & Isom, D. G. Predictable cholesterol binding sites in GPCRs lack consensus motifs. Structure 29, 499–506.e3 (2021).
Huang, S. K. et al. Allosteric modulation of the adenosine A2A receptor by cholesterol. eLife 11, e73901 (2022).
Li, L. B., Vorobyov, I. & Allen, T. W. The role of membrane thickness in charged protein–lipid interactions. Biochim. Biophys. Acta - Biomembr. 1818, 135–145 (2012).
Mondal, S. et al. Membrane driven spatial organization of GPCRs. Sci Rep. 3, 2909 (2013).
Róg, T. & Vattulainen, I. Cholesterol, sphingolipids, and glycolipids: what do we know about their role in raft-like membranes? Chem. Phys. Lipids 184, 82–104 (2014).
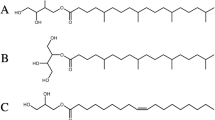
Kulig, W. et al. How well does cholesteryl hemisuccinate mimic cholesterol in saturated phospholipid bilayers? J. Mol. Model. 20, 2121 (2014).
Warne, T. et al. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469, 241–244 (2011).
Cang, X. et al. Cholesterol-β1AR interaction versus cholesterol-β2AR interaction. Proteins Struct. Funct. Bioinf. 82, 760–770 (2014).
Quillin, M. L., Breyer, W. A., Griswold, I. J. & Matthews, B. W. Size versus polarizability in protein-ligand interactions: binding of noble gases within engineered cavities in phage T4 lysozyme. J. Mol. Biol. 302, 955–977 (2000).
Guixà-González, R. et al. Membrane cholesterol access into a G-protein-coupled receptor. Nat. Commun. 8, 14505 (2017).
Warne, T., Edwards, P. C., Doré, A. S., Leslie, A. G. W. & Tate, C. G. Molecular basis for high-affinity agonist binding in GPCRs. Science 364, 775 (2019).
Liu, X. et al. An allosteric modulator binds to a conformational hub in the β2 adrenergic receptor. Nat. Chem. Biol. 16, 749–755 (2020).
Lu, J. et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 24, 570–577 (2017).
Zhang, D. et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 520, 317–321 (2015).
Vial, C., Fung, C. Y. E., Goodall, A. H., Mahaut-Smith, M. P. & Evans, R. J. Differential sensitivity of human platelet P2X1 and P2Y1 receptors to disruption of lipid rafts. Biochem. Biophys. Res. Commun. 343, 415–419 (2006).
Prangé, T. et al. Exploring hydrophobic sites in proteins with xenon or krypton. Proteins Struct. Funct. Bioinf. 30, 61–73 (1998).
Paila, Y. D., Jindal, E., Goswami, S. K. & Chattopadhyay, A. Cholesterol depletion enhances adrenergic signaling in cardiac myocytes. Biochim. Biophys. Acta - Biomembr. 1808, 461–465 (2011).
Yeliseev, A. et al. Cholesterol as a modulator of cannabinoid receptor CB2 signaling. Sci Rep. 11, 3706 (2021).
Xiao, P. et al. Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell 184, 943–956.e18 (2021).
Robertson, N. et al. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 553, 111–114 (2018).
Cheng, R. K. Y. et al. Structural insight into allosteric modulation of protease-activated receptor 2. Nature 545, 112–115 (2017).
Thal, D. M., Glukhova, A., Sexton, P. M. & Christopoulos, A. Structural insights into G-protein-coupled receptor allostery. Nature 559, 45–53 (2018).
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012).
Rasmussen, S. G. F. et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011).
Warne, T., Serrano-Vega, M. J., Tate, C. G. & Schertler, G. F. X. Development and crystallization of a minimal thermostabilised G protein-coupled receptor. Protein Expr. Purif. 65, 204–213 (2009).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010).
Thorn, A. & Sheldrick, G. M. ANODE: anomalous and heavy-atom density calculation. J. Appl. Crystallogr. 44, 1285–1287 (2011).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
The PyMOL Molecular Graphics System, v.1.8 (Schrödinger, 2015).
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008).
Sklenář, V. & Bax, A. Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J. Magn. Reson. 74, 469–479 (1987).
Scheuermann, T. H. & Brautigam, C. A. High-precision, automated integration of multiple isothermal titration calorimetric thermograms: new features of NITPIC. Methods 76, 87–98 (2015).
Zhao, H., Piszczek, G. & Schuck, P. SEDPHAT–a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148 (2015).
Brautigam, C. A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015).
Acknowledgements
This work was supported by the Swiss National Science Foundation (Grants CRSK-3_195592 to L.A.A. and 31-149927, 31-173089 and 31-201270 to S.G.). We gratefully acknowledge M. Schaffhauser, P. Schlenker and R. Strittmatter (Biozentrum Central Mechanical Workshop) as well as S. Saner (Biozentrum Central Electronics Workshop) for designing and building the xenon pressure chamber apparatus, the Paul Scherrer Institut, Villigen, Switzerland for synchrotron radiation beamtime at beamline PXIII, the Biozentrum Biophysics Facility for access to their instruments, J. Steyaert for the generous gift of the Nb80 plasmid, as well as H.-J. Sass, C. Tate, T. Maier and T. Schirmer for helpful discussions.
Author information
Authors and Affiliations
Contributions
L.A.A. and S.G. conceived the study. L.A.A. and A.G. expressed and purified proteins and recorded NMR spectra. L.A.A. recorded high-pressure NMR experiments. L.A.A. analysed and interpreted the NMR data. R.D.T. and S.E. recorded the X-ray data. R.D.T. analysed the X-ray data. T.M. and T.S. performed and analysed ITC experiments. L.A.A., R.D.T. and S.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and Tables 1–3.
Supplementary Table 1
Numerical source of 1H NMR resonance intensities plotted in Supplementary Fig. 3b.
Source data
Source Data Fig. 3
Numerical Source Data for Fig. 3c.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abiko, L.A., Dias Teixeira, R., Engilberge, S. et al. Filling of a water-free void explains the allosteric regulation of the β1-adrenergic receptor by cholesterol. Nat. Chem. 14, 1133–1141 (2022). https://doi.org/10.1038/s41557-022-01009-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-022-01009-9
This article is cited by
-
Binding kinetics drive G protein subtype selectivity at the β1-adrenergic receptor
Nature Communications (2024)