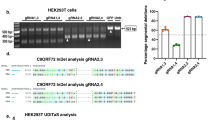
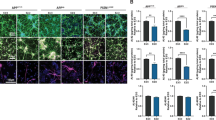
Abstract
The pathology of familial Alzheimer’s disease, which is caused by dominant mutations in the gene that encodes amyloid-beta precursor protein (APP) and in those that encode presenilin 1 and presenilin 2, is characterized by extracellular amyloid plaques and intracellular neurofibrillary tangles in multiple brain regions. Here we show that the brain-wide selective disruption of a mutated APP allele in transgenic mouse models carrying the human APP Swedish mutation alleviates amyloid-beta-associated pathologies for at least six months after a single intrahippocampal administration of an adeno-associated virus that encodes both Cas9 and a single-guide RNA that targets the mutation. We also show that the deposition of amyloid-beta, as well as microgliosis, neurite dystrophy and the impairment of cognitive performance, can all be ameliorated when the CRISPR–Cas9 construct is delivered intravenously via a modified adeno-associated virus that can cross the blood–brain barrier. Brain-wide disease-modifying genome editing could represent a viable strategy for the treatment of familial Alzheimer’s disease and other monogenic diseases that affect multiple brain regions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw data from whole-genome sequencing have been deposited in the NCBI Sequence Read Archive (SRA), with accession code PRJNA733582. The other raw and analysed datasets generated during the study are available for research purposes from the corresponding author on reasonable request, as they are too large to be publicly shared.
References
Cai, Y., An, S. S. & Kim, S. Mutations in presenilin 2 and its implications in Alzheimer’s disease and other dementia-associated disorders. Clin. Interv. Aging 10, 1163–1172 (2015).
Campion, D. et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am. J. Hum. Genet. 65, 664–670 (1999).
Cacace, R., Sleegers, K. & Van Broeckhoven, C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 12, 733–748 (2016).
Harper, P. S. The epidemiology of Huntington’s disease. Hum. Genet. 89, 365–376 (1992).
Chio, A. et al. Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology 41, 118–130 (2013).
Selkoe, D. J. & Hardy, J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Serrano-Pozo, A., Frosch, M. P., Masliah, E. & Hyman, B. T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 1, a006189 (2011).
Braak, H. & Braak, E. Frequency of stages of Alzheimer-related lesions in different age categories. Neurobiol. Aging 18, 351–357 (1997).
Ingusci, S., Verlengia, G., Soukupova, M., Zucchini, S. & Simonato, M. Gene therapy tools for brain diseases. Front. Pharmacol. 10, 724 (2019).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Deverman, B. E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016).
Bedbrook, C. N., Deverman, B. E. & Gradinaru, V. Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci. 41, 323–348 (2018).
Zeng, J. et al. TRIM9-mediated resolution of neuroinflammation confers neuroprotection upon ischemic stroke in mice. Cell Rep. 27, 549–560.e6 (2019).
Ortiz-Virumbrales, M. et al. CRISPR/Cas9-correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 N141I neurons. Acta Neuropathol. Commun. 5, 77 (2017).
Gyorgy, B. et al. CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol. Ther. Nucleic Acids 11, 429–440 (2018).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Wang, B. et al. γ-Secretase gene mutations in familial acne inversa. Science 330, 1065 (2010).
Mullan, M. et al. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of β-amyloid. Nat. Genet. 1, 345–347 (1992).
Citron, M. et al. Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proc. Natl Acad. Sci. USA 91, 11993–11997 (1994).
Johnston, J. A. et al. Increased β-amyloid release and levels of amyloid precursor protein (APP) in fibroblast cell lines from family members with the Swedish Alzheimer’s disease APP670/671 mutation. FEBS Lett. 354, 274–278 (1994).
Wiedenheft, B. et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl Acad. Sci. USA 108, 10092–10097 (2011).
Gao, X. et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221 (2018).
Mashiko, D. et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3, 3355 (2013).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Kimura, R. & Ohno, M. Impairments in remote memory stabilization precede hippocampal synaptic and cognitive failures in 5XFAD Alzheimer mouse model. Neurobiol. Dis. 33, 229–235 (2009).
Shao, C. Y., Mirra, S. S., Sait, H. B., Sacktor, T. C. & Sigurdsson, E. M. Postsynaptic degeneration as revealed by PSD-95 reduction occurs after advanced Aβ and tau pathology in transgenic mouse models of Alzheimer’s disease. Acta Neuropathol. 122, 285–292 (2011).
Gowrishankar, S. et al. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl Acad. Sci. USA 112, E3699–E3708 (2015).
Sharoar, M. G., Hu, X., Ma, X. M., Zhu, X. & Yan, R. Sequential formation of different layers of dystrophic neurites in Alzheimer’s brains. Mol. Psychiatry 24, 1369–1382 (2019).
Spires-Jones, T. L. & Hyman, B. T. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron 82, 756–771 (2014).
Woodhouse, A., West, A. K., Chuckowree, J. A., Vickers, J. C. & Dickson, T. C. Does β-amyloid plaque formation cause structural injury to neuronal processes? Neurotox. Res. 7, 5–15 (2005).
Jankowsky, J. L. et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet. 13, 159–170 (2004).
Garcia-Alloza, M. et al. Characterization of amyloid deposition in the APPswe/PS1dE9 mouse model of Alzheimer disease. Neurobiol. Dis. 24, 516–524 (2006).
Fu, A. K. et al. IL-33 ameliorates Alzheimer’s disease-like pathology and cognitive decline. Proc. Natl Acad. Sci. USA 113, E2705–E2713 (2016).
Lalonde, R., Kim, H. D., Maxwell, J. A. & Fukuchi, K. Exploratory activity and spatial learning in 12-month-old APP695SWE/co+PS1/ΔE9 mice with amyloid plaques. Neurosci. Lett. 390, 87–92 (2005).
Choi, J. H. et al. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain 7, 17 (2014).
Patricio, M. I., Barnard, A. R., Orlans, H. O., McClements, M. E. & MacLaren, R. E. Inclusion of the woodchuck hepatitis virus posttranscriptional regulatory element enhances AAV2-driven transduction of mouse and human retina. Mol. Ther. Nucleic Acids 6, 198–208 (2017).
Jawhar, S., Trawicka, A., Jenneckens, C., Bayer, T. A. & Wirths, O. Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Aβ aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 33, 196.e29–40 (2012).
Tible, M. et al. PKR knockout in the 5xFAD model of Alzheimer’s disease reveals beneficial effects on spatial memory and brain lesions. Aging Cell 18, e12887 (2019).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Cruts, M., Theuns, J. & Van Broeckhoven, C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 33, 1340–1344 (2012).
Kleinstiver, B. P. et al. Broadening the targeting range of Staphylococcus aureus CRISPR–Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298 (2015).
Nagata, K. et al. Generation of App knock-in mice reveals deletion mutations protective against Alzheimer’s disease-like pathology. Nat. Commun. 9, 1800 (2018).
Park, H. et al. In vivo neuronal gene editing via CRISPR–Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 22, 524–528 (2019).
Filser, S. et al. Pharmacological inhibition of BACE1 impairs synaptic plasticity and cognitive functions. Biol. Psychiatry 77, 729–739 (2015).
Ou-Yang, M.-H. et al. Axonal organization defects in the hippocampus of adult conditional BACE1 knockout mice. Sci. Transl. Med. 10, eaao5620 (2018).
Blume, T. et al. BACE1 inhibitor MK-8931 alters formation but not stability of dendritic spines. Front. Aging Neurosci. 10, 229 (2018).
Ravindra Kumar, S. et al. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods 17, 541–550 (2020).
Flytzanis, N. C. et al. Broad gene expression throughout the mouse and marmoset brain after intravenous delivery of engineered AAV capsids. Preprint at bioRxiv https://doi.org/10.1101/2020.06.16.152975 (2020).
Bloch, D. B. et al. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol. Cell. Biol. 20, 6138–6146 (2000).
Mo, W. & Zhang, J. T. Human ABCG2: structure, function, and its role in multidrug resistance. Int J. Biochem Mol. Biol. 3, 1–27 (2012).
Liang, Z. et al. The pseudokinase CaMKv is required for the activity-dependent maintenance of dendritic spines. Nat. Commun. 7, 13282 (2016).
Sun, J. et al. CRISPR/Cas9 editing of APP C-terminus attenuates β-cleavage and promotes α-cleavage. Nat. Commun. 10, 53 (2019).
Hocquemiller, M., Giersch, L., Audrain, M., Parker, S. & Cartier, N. Adeno-associated virus-based gene therapy for CNS diseases. Hum. Gene Ther. 27, 478–496 (2016).
Hudry, E. & Vandenberghe, L. H. Therapeutic AAV gene transfer to the nervous system: a clinical reality. Neuron 101, 839–862 (2019).
Challis, R. C. et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 14, 379–414 (2019).
Keane, T. M. et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011).
Wojtowicz, J. M. & Kee, N. BrdU assay for neurogenesis in rodents. Nat. Protoc. 1, 1399–1405 (2006).
Styren, S. D., Hamilton, R. L., Styren, G. C. & Klunk, W. E. X-34, a fluorescent derivative of Congo red: a novel histochemical stain for Alzheimer’s disease pathology. J. Histochem. Cytochem. 48, 1223–1232 (2000).
Seo, J. et al. Activity-dependent p25 generation regulates synaptic plasticity and Aβ-induced cognitive impairment. Cell 157, 486–498 (2014).
Lee, K. et al. Replenishment of microRNA-188-5p restores the synaptic and cognitive deficits in 5XFAD mouse model of Alzheimer’s disease. Sci. Rep. 6, 34433 (2016).
Devi, L. & Ohno, M. Mitochondrial dysfunction and accumulation of the β-secretase-cleaved C-terminal fragment of APP in Alzheimer’s disease transgenic mice. Neurobiol. Dis. 45, 417–424 (2012).
Acknowledgements
We thank K. Liu, W.-Y. Fu, X. Wang, K.-W. Hung, C. Kwong, R. M. Delos Reyes, K. Cheung, N. Mullapudi, E. Tam, J. Zhang, H. Cao, S.-F. Li and P.-O. Chiu of the Hong Kong University of Science and Technology; and B. E. Deverman, K. Beadle and Y. Lei of California Institute of Technology for their excellent technical assistance. We are grateful to all members of the Ip laboratory for discussions. This study was supported in part by the National Key R&D Program of China (2018YFE0203600), the Guangdong Provincial Key S&T Program (2018B030336001), the Guangdong Provincial Fund for Basic and Applied Basic Research (2019B1515130004), the Hong Kong Research Grants Council Theme-based Research Scheme (T13-605/18-W), the Area of Excellence Scheme of the University Grants Committee (AoE/M-604/16), the Innovation and Technology Commission (ITCPD/17-9), the Lee Hysan Foundation (LHF17SC01), the Shenzhen Knowledge Innovation Program (JCYJ20180507183642005 and JCYJ20200109115631248), the Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (project number: 2019SHIBS0001), the HKUST-SIAT Joint Laboratory for Brain Science for technical platform support, and the Beckman Institute for CLARITY, Optogenetics and Vector Engineering Research for technology development and broad dissemination (clover.caltech.edu (V.G.) and the CZI Neurodegeneration Challenge Network (V.G.)).
Author information
Authors and Affiliations
Contributions
Y.D., T.Y., A.K.Y.F. and N.Y.I. designed the research; Y.D., T.Y., Z.Q., A.M., X.Z., K.-C.L., Yuewen Chen and Yu Chen performed the research; Y.D., T.Y., A.K.Y.F. and N.Y.I. analysed the data; X.Z. performed bioinformatics analysis; V.G. and N.Y.I. contributed to the design and availability of reagents/analytic tools; and Y.D., T.Y., A.K.Y.F. and N.Y.I. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks Brandon Harvey, Brigitte van Zundert and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures and tables.
Supplementary Dataset
Identified somatic mutations as potential off-target events.
Rights and permissions
About this article
Cite this article
Duan, Y., Ye, T., Qu, Z. et al. Brain-wide Cas9-mediated cleavage of a gene causing familial Alzheimer’s disease alleviates amyloid-related pathologies in mice. Nat Biomed Eng 6, 168–180 (2022). https://doi.org/10.1038/s41551-021-00759-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00759-0
This article is cited by
-
Whole-brain in vivo base editing reverses behavioral changes in Mef2c-mutant mice
Nature Neuroscience (2024)
-
A year in review: brain barriers and brain fluids research in 2022
Fluids and Barriers of the CNS (2023)
-
Removal of a partial genomic duplication restores synaptic transmission and behavior in the MyosinVA mutant mouse Flailer
BMC Biology (2023)
-
Cas9-mediated replacement of expanded CAG repeats in a pig model of Huntington’s disease
Nature Biomedical Engineering (2023)
-
Non-viral approaches for gene therapy and therapeutic genome editing across the blood–brain barrier
Med-X (2023)