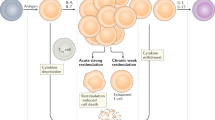
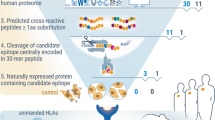
Abstract
Adaptive immune responses and immunopathogeneses are based on the ability of T cells to respond to specific antigens. Consequently, understanding T-cell recognition patterns in health and disease involves studying the complexity and genetic heterogeneity of the antigen recognition pathway, which includes both T-cell receptors and the antigen-presentation machinery. In this Perspective, we overview the development and use of technologies for assessing T-cell recognition in a clinical context, and discuss how knowledge of T-cell recognition pathways can be critical before, during and after disease treatment. The ability to assess T-cell-mediated immunity in individual patients during disease progression might enable the identification of patient-specific biomarkers that predict therapeutic efficacy and response. Effective strategies for the complex analysis of T-cell specificity in clinical settings are highly desirable and could complement current approaches for the monitoring of therapy responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
02 January 2018
In the version of this Perspective originally published, in Fig. 4, in the schematic for the DNA barcoded multimers, the barcodes were missing; they have now been included and the figure updated in all versions of the Perspective.
References
Chien, Y. H. & Davis, M. M. How alpha beta T-cell receptors ‘see’ peptide/MHC complexes. Immunol. Today 14, 597–602 (1993).
Germain, R. N. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell 76, 287–299 (1994).
Pape, K. A. et al. Use of adoptive transfer of T-cell-antigen-receptor-transgenic T cell for the study of T-cell activation in vivo. Immunol. Rev. 156, 67–78 (1997).
Hogquist, K. A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
Robey, E. A. et al. The level of CD8 expression can determine the outcome of thymic selection. Cell 69, 1089–1096 (1992).
Akram, A. & Inman, R. D. Immunodominance: a pivotal principle in host response to viral infections. Clin. Immunol. 143, 99–115 (2012).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Davis, M. M., Altman, J. D. & Newell, E. W. Interrogating the repertoire: broadening the scope of peptide-MHC multimer analysis. Nat. Rev. Immunol. 11, 551–558 (2011).
Bentzen, A. K. & Hadrup, S. R. Evolution of MHC-based technologies used for detection of antigen-responsive T cells. Cancer Immunol. Immunother. 66, 657–666 (2017).
Robbins, P. F. et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 29, 917–924 (2011).
Morgan, R. A. et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36, 133–151 (2013).
Neuenhahn, M. et al. Transfer of minimally manipulated CMV-specific T cells from stem cell or third-party donors to treat CMV infection after allo-HSCT. Leukemia https://doi.org/10.1038/leu.2017.16 (2017).
Cobbold, M. et al. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. J. Exp. Med. 202, 379–386 (2005).
Wambre, E. et al. Specific immunotherapy modifies allergen-specific CD4+ T-cell responses in an epitope-dependent manner. J. Allergy Clin. Immunol. 133, 872–879.e7 (2014).
Odegard, J. M., Nepom, G. T. & Wambre, E. Biomarkers for antigen immunotherapy in allergy and type 1 diabetes. Clin. Immunol. 161, 44–50 (2015).
Radvanyi, L. G. et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin. Cancer Res. 18, 6758–6770 (2012).
Darrah, P. A. et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13, 843–850 (2007).
Lee, P. P. et al. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat. Med. 5, 677–685 (1999).
Coulie, P. G., Van den Eynde, B. J., van der Bruggen, P. & Boon, T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014).
Coulie, P. G. et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J. Exp. Med. 180, 35–42 (1994).
Van Nuffel, A. M. T. et al. Intravenous and intradermal TriMix-dendritic cell therapy results in a broad T-cell response and durable tumor response in a chemorefractory stage IV-M1c melanoma patient. Cancer Immunol. Immunother. 61, 1033–1043 (2012).
Andersen, R. S. et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 72, 1642–1650 (2012).
Kvistborg, P. et al. TIL therapy broadens the tumor-reactive CD8+ T cell compartment in melanoma patients. Oncoimmunology 1, 409–418 (2012).
Rizvi, N. A. et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2014).
Van Allen, E. M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Strønen, E. et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352, 1337–1341 (2016).
Gros, A. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438 (2016).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016).
van Rooij, N. et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442 (2013).
Bassani-Sternberg, M. et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016).
Bentzen, A. K. et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 34, 1037–1045 (2016).
Wick, D. A. et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin. Cancer Res. 20, 1125–1134 (2014).
Rajasagi, M. et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 124, 453–462 (2014).
Verdegaal, E. M. E. et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 536, 91–95 (2016).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Gubin, M. M. et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014).
Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Palmer, D. C. et al. Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J. Exp. Med. 212, 2095–2113 (2015).
Chidrawar, S. et al. Cytomegalovirus-seropositivity has a profound influence on the magnitude of major lymphoid subsets within healthy individuals. Clin. Exp. Immunol. 155, 423–432 (2009).
Gordon, C. L. et al. Tissue reservoirs of antiviral T cell immunity in persistent human CMV infection. J. Exp. Med. 214, 651–667 (2017).
Colugnati, F. A. B., Staras, S. A. S., Dollard, S. C. & Cannon, M. J. Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect. Dis. 7, 71 (2007).
Hadrup, S. R. et al. Longitudinal studies of clonally expanded CD8 T cells reveal a repertoire shrinkage predicting mortality and an increased number of dysfunctional cytomegalovirus-specific T cells in the very elderly. J. Immunol. 176, 2645–2653 (2006).
Furman, D. et al. Cytomegalovirus infection enhances the immune response to influenza. Sci. Transl. Med. 7, 281ra43 (2015).
Kamphorst, A. O. et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427 (2017).
Williams, M. A. et al. Cutting edge: persistent viral infection prevents tolerance induction and escapes immune control following CD28/CD40 blockade-based regimen. J. Immunol. 169, 5387–5391 (2002).
Lanzavecchia, A. & Sallusto, F. Understanding the generation and function of memory T cell subsets. Curr. Opin. Immunol. 17, 326–332 (2005).
Sylwester, A. W. et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685 (2005).
Lindestam Arlehamn, C. S. et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected South Africans. PLoS Pathog. 12, e1005760 (2016).
Lindestam Arlehamn, C. S., Lewinsohn, D., Sette, A. & Lewinsohn, D. Antigens for CD4 and CD8 T cells in tuberculosis. Cold Spring Harb. Perspect. Med. 4, a018465 (2014).
Höhn, H. et al. MHC class II tetramer guided detection of Mycobacterium tuberculosis-specific CD4+ T cells in peripheral blood from patients with pulmonary tuberculosis. Scand. J. Immunol. 65, 467–478 (2007).
Newell, E. W. et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat. Biotechnol. 31, 623–629 (2013).
Kracht, M. J. L. et al. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat. Med. 23, 501–507 (2017).
Roep, B. O., Kracht, M. J., van Lummel, M. & Zaldumbide, A. A roadmap of the generation of neoantigens as targets of the immune system in type 1 diabetes. Curr. Opin. Immunol. 43, 67–73 (2016).
Salou, M., Nicol, B., Garcia, A. & Laplaud, D.-A. Involvement of CD8+ T cells in multiple sclerosis. Front. Immunol. 6, 604 (2015).
McGinty, J. W. et al. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes 63, 3033–3040 (2014).
Rondas, D. et al. Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes 64, 573–586 (2015).
McLaughlin, R. J., Spindler, M. P., van Lummel, M. & Roep, B. O. Where, how, and when: positioning posttranslational modification within type 1 diabetes pathogenesis. Curr. Diab. Rep. 16, 63 (2016).
Suárez-Fueyo, A., Bradley, S. J. & Tsokos, G. C. T cells in systemic Lupus Erythematosus. Curr. Opin. Immunol. 43, 32–38 (2016).
Carvalheiro, H., da Silva, J. A. P. & Souto-Carneiro, M. M. Potential roles for CD8+ T cells in rheumatoid arthritis. Autoimmun. Rev. 12, 401–409 (2013).
Andersen, R. S. et al. High frequency of T cells specific for cryptic epitopes in melanoma patients. Oncoimmunology 2, e25374 (2013).
Spath, S. et al. Dysregulation of the cytokine GM-CSF induces spontaneous phagocyte invasion and immunopathology in the central nervous system. Immunity 46, 245–260 (2017).
Yang, J. et al. Expression of HLA-DP0401 molecules for identification of DP0401 restricted antigen specific T cells. J. Clin. Immunol. 25, 428–436 (2005).
Archila, L. L. D. & Kwok, W. W. Tetramer-guided epitope mapping: a rapid approach to identify HLA-restricted T-cell epitopes from composite allergens. Methods Mol. Biol. 1592, 199–209 (2017).
Hinz, D. et al. Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen-specific modulation of immune reactivity. Clin. Exp. Allergy 46, 705–719 (2016).
Wambre, E., James, E. A. & Kwok, W. W. Characterization of CD4+ T cell subsets in allergy. Curr. Opin. Immunol. 24, 700–706 (2012).
Lu, Y.-C. et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Cancer Res. 20, 3401–3410 (2014).
Birnbaum, M. E., Dong, S. & Garcia, K. C. Diversity-oriented approaches for interrogating T-cell receptor repertoire, ligand recognition, and function. Immunol. Rev. 250, 82–101 (2012).
Birnbaum, M. E. et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087 (2014).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Newell, E. W. Higher throughput methods of identifying T cell epitopes for studying outcomes of altered antigen processing and presentation. Front. Immunol. 4, 430 (2013).
Vigneron, N. et al. An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004).
Liepe, J. et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016).
Schumacher, T. N. M. et al. Peptide selection by MHC class I molecules. Nature 350, 703–706 (1991).
Nielsen, M., Justesen, S., Lund, O., Lundegaard, C. & Buus, S. NetMHCIIpan-2.0—Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res. 6, 9 (2010).
Andreatta, M. et al. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics 67, 641–650 (2015).
Braendstrup, P. et al. MHC class II tetramers made from isolated recombinant α and β chains refolded with affinity-tagged peptides. PLoS ONE 8, e73648 (2013).
Crawford, F., Kozono, H., White, J., Marrack, P. & Kappler, J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 8, 675–682 (1998).
Rahim, A. et al. Potent T cell activation with dimeric peptide–major histocompatibility complex class II ligand: the role of CD4 coreceptor. J. Exp. Med. 188, 1633–1640 (1998).
Toebes, M. et al. Design and use of conditional MHC class I ligands. Nat. Med. 12, 246–251 (2006).
Saini, S. K. et al. Dipeptides catalyze rapid peptide exchange on MHC class I molecules. Proc. Natl Acad. Sci. USA 112, 202–207 (2015).
Leisner, C. et al. One-pot, mix-and-read peptide-MHC tetramers. PLoS ONE 3, e1678 (2008).
Day, C. L. et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest. 112, 831–842 (2003).
Landais, E. et al. New design of MHC class II tetramers to accommodate fundamental principles of antigen presentation. J. Immunol. 183, 7949–7957 (2009).
Hadrup, S. R. et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods 6, 520–526 (2009).
Newell, E. W., Klein, L. O., Yu, W. & Davis, M. M. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat. Methods 6, 497–499 (2009).
Kvistborg, P. et al. Thinking outside the gate: single-cell assessments in multiple dimensions. Immunity 42, 591–592 (2015).
Appay, V., Van Lier, R. A. W., Sallusto, F. & Roederer, M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry Part A 73, 975–983 (2008).
Ornatsky, O. I., Baranov, V. I., Bandura, D. R., Tanner, S. D. & Dick, J. Messenger RNA detection in leukemia cell lines by novel metal-tagged in situ hybridization using inductively coupled plasma mass spectrometry. Transl. Oncogenomics 1, 1–9 (2006).
Bandura, D. R. et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81, 6813–6822 (2009).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Cheng, Y. & Newell, E. W. Deep profiling human T cell heterogeneity by mass cytometry. Adv. Immunol. 131, 101–134 (2016).
Mair, F. et al. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur. J. Immunol. 46, 34–43 (2016).
Newell, E. W. & Cheng, Y. Mass cytometry: blessed with the curse of dimensionality. Nat. Immunol. 17, 890–895 (2016).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Wong, M. T. et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45, 442–456 (2015).
Newell, E. W., Sigal, N., Bendall, S. C., Nolan, G. P. & Davis, M. M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36, 142–152 (2012).
Cheng, Y., Wong, M. T., van der Maaten, L. & Newell, E. W. Categorical analysis of human T cell heterogeneity with one-dimensional soli-expression by nonlinear stochastic embedding. J. Immunol. 196, 924–932 (2016).
Wistuba-Hamprecht, K. et al. Establishing high dimensional immune signatures from peripheral blood via mass cytometry in a discovery cohort of stage IV melanoma patients. J. Immunol. 198, 927–936 (2017).
Krams, S. M., Schaffert, S., Lau, A. H. & Martinez, O. M. Applying mass cytometry to the analysis of lymphoid populations in transplantation. Am. J. Transplant. 17, 1992–1999 (2017).
Brooks, M. Insulinoma and abdominal tuberculosis. Scott. Med. J. 33, 207–208 (1988).
Xu, Q., Schlabach, M. R., Hannon, G. J. & Elledge, S. J. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl Acad. Sci. USA 106, 2289–2294 (2009).
MacBeath, G. & Schreiber, S. L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000).
Soen, Y., Chen, D. S., Kraft, D. L., Davis, M. M. & Brown, P. O. Detection and characterization of cellular immune responses using peptide-MHC microarrays. PLoS Biol. 1, e65 (2003).
Stone, J. D., Demkowicz, W. E. & Stern, L. J. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proc. Natl Acad. Sci. USA 102, 3744–3749 (2005).
Chen, D. S. et al. Marked differences in human melanoma antigen-specific T cell responsiveness after vaccination using a functional microarray. PLoS Med. 2, e265 (2005).
Deviren, G., Gupta, K., Paulaitis, M. E. & Schneck, J. P. Detection of antigen-specific T cells on p/MHC microarrays. J. Mol. Recognit. 20, 32–38 (2007).
Kwong, G. A. et al. Modular nucleic acid assembled p/MHC microarrays for multiplexed sorting of antigen-specific T cells. J. Am. Chem. Soc. 131, 9695–9703 (2009).
Brooks, S. E. et al. Application of the pMHC array to characterise tumour antigen specific T cell populations in leukaemia patients at disease diagnosis. PLoS ONE 10, e0140483 (2015).
Klinger, M. et al. Multiplex identification of antigen-specific T cell receptors using a combination of immune assays and immune receptor sequencing. PLoS ONE 10, e0141561 (2015).
Novak, E. J. et al. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J. Immunol. 166, 6665–6670 (2001).
Robins, H. S. et al. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood 114, 4099–4107 (2009).
Davis, M. M. & Bjorkman, P. J. T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988).
van Buuren, M. M. et al. HLA micropolymorphisms strongly affect peptide-MHC multimer-based monitoring of antigen-specific CD8+ T cell responses. J. Immunol. 192, 641–648 (2014).
Frøsig, T. M. et al. Design and validation of conditional ligands for HLA-B*08:01, HLA-B*15:01, HLA-B*35:01, and HLA-B*44:05. Cytom. Part A 87, 967–975 (2015).
Mason, D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19, 395–404 (1998).
Márquez, A. C. & Horwitz, M. S. The role of latently infected B cells in CNS autoimmunity. Front. Immunol. 6, 544 (2015).
Lundegaard, C., Lund, O., Buus, S. & Nielsen, M. Major histocompatibility complex class I binding predictions as a tool in epitope discovery. Immunology 130, 309–318 (2010).
Abelin, J. G. et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017).
Malaker, S. A. et al. Identification and characterization of complex glycosylated peptides presented by the MHC class II processing pathway in melanoma. J. Proteome Res. 16, 228–237 (2017).
Fritsch, E. F. et al. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol. Res. 2, 522–529 (2014).
The problem with neoantigen prediction. Nat. Biotechnol. 35, 97 (2017).
Liu, X. S. & Mardis, E. R. Applications of immunogenomics to cancer. Cell 168, 600–612 (2017).
Osborne, G. W., Andersen, S. B. & Battye, F. L. Development of a novel cell sorting method that samples population diversity in flow cytometry. Cytometry. A 87, 1047–1051 (2015).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
van Buggenum, J. A. G. L. et al. A covalent and cleavable antibody-DNA conjugation strategy for sensitive protein detection via immuno-PCR. Sci. Rep. 6, 22675 (2016).
Moon, J. J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Yu, W. et al. Clonal deletion prunes but does not eliminate self-specific αβ CD8+ T lymphocytes. Immunity 42, 929–941 (2015).
Zoete, V., Irving, M., Ferber, M., Cuendet, M. A. & Michielin, O. Structure-based, rational design of T cell receptors. Front. Immunol. 4, 268 (2013).
Dash, P. et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017).
Glanville, J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017).
Sela-culang, I. et al. Resource using a combined computational-experimental approach to predict antibody-specific B cell epitopes. Struct. Des. 22, 646–657 (2014).
Furman, D. et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat. Med. 23, 174–184 (2017).
Sette, A. & Peters, B. Immune epitope mapping in the post-genomic era: lessons for vaccine development. Curr. Opin. Immunol. 19, 106–110 (2007).
Anderson, R. P. & Jabri, B. Vaccine against autoimmune disease: antigen-specific immunotherapy. Curr. Opin. Immunol. 25, 410–417 (2013).
Czerkinsky, C. C., Nilsson, L. A., Nygren, H., Ouchterlony, O. & Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J. Immunol. Methods 65, 109–121 (1983).
Draenert, R. et al. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275, 19–29 (2003).
Waldrop, S. L., Pitcher, C. J., Peterson, D. M., Maino, V. C. & Picker, L. J. Determination of antigen-specific memory/effector CD4+ T cell frequencies by flow cytometry: evidence for a novel, antigen-specific homeostatic mechanism in HIV-associated immunodeficiency. J. Clin. Invest. 99, 1739–1750 (1997).
Bacher, P. et al. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J. Immunol. 190, 3967–3976 (2013).
Bacher, P. et al. Regulatory T cell specificity directs tolerance versus allergy against aeroantigens in humans. Cell 167, 1067–1078e16 (2016).
Appay, V. & Rowland-Jones, S. L. The assessment of antigen-specific CD8+ T cells through the combination of MHC class I tetramer and intracellular staining. J. Immunol. Methods 268, 9–19 (2002).
Geiger, R., Duhen, T., Lanzavecchia, A. & Sallusto, F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J. Exp. Med. 206, 1525–1534 (2009).
Becattini, S. et al. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science 347, 400–406 (2015).
Cox, A. L. et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science 264, 716–719 (1994).
Robbins, P. D. & Morelli, A. E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208 (2014).
Amir, E. D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Kozono, H., White, J., Clements, J., Marrack, P. & Kappler, J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature 369, 151–154 (1994).
Bankovich, A. J., Girvin, A. T., Moesta, A. K. & Garcia, K. C. Peptide register shifting within the MHC groove: theory becomes reality. Mol. Immunol. 40, 1033–1039 (2004).
Lin, H. H., Zhang, G. L., Tongchusak, S., Reinherz, E. L. & Brusic, V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics 9(Suppl. 12), S22 (2008).
Su, L. F., Kidd, B. A., Han, A., Kotzin, J. J. & Davis, M. M. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity 38, 373–383 (2013).
Uchtenhagen, H. et al. Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining. Nat. Commun. 7, 12614 (2016).
Lissina, A. et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J. Immunol. Methods 340, 11–24 (2009).
Xie, J. et al. Photocrosslinkable pMHC monomers stain T cells specifically and cause ligand-bound TCRs to be ‘preferentially’ transported to the cSMAC. Nat. Immunol. 13, 674–680 (2012).
Acknowledgements
The work was funded by the European Research Council (ERC), ERC starting grant and the Lundbeck Foundation fellowship (S.R.H.) and the Singapore Immunology Network (SIgN) (E.W.N.).
Author information
Authors and Affiliations
Contributions
S.R.H. and E.W.N. conceived and wrote this Perspective.
Corresponding author
Ethics declarations
Competing interests
E.W.N. is a board director and shareholder of immunoSCAPE Pte. Ltd. S.R.H. is a co-founder of Immumap Services.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A correction to this article is available online at https://doi.org/10.1038/s41551-017-0176-8.
Rights and permissions
About this article
Cite this article
Hadrup, S.R., Newell, E. Determining T-cell specificity to understand and treat disease. Nat Biomed Eng 1, 784–795 (2017). https://doi.org/10.1038/s41551-017-0143-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-017-0143-4
This article is cited by
-
Nanoscale organization of two-dimensional multimeric pMHC reagents with DNA origami for CD8+ T cell detection
Nature Communications (2022)