Abstract
The generation of molecules with artificial intelligence (AI) or, more specifically, machine learning (ML), is poised to revolutionize materials discovery. Potential applications range from development of potent drugs to efficient carbon capture and separation technologies. However, existing computational discovery frameworks for polymer membranes lack automated training data creation, generative design, and physical performance validation at meso-scale where complex properties of amorphous materials emerge. The methodological gaps are less relevant to the ML design of individual molecules such as the monomers which constitute the building blocks of polymers. Here, we report automated discovery of complex materials through inverse molecular design which is informed by meso-scale target features and process figures-of-merit. We have explored the multi-scale discovery regime by computationally generating and validating hundreds of polymer candidates designed for application in post-combustion carbon dioxide filtration. Specifically, we have validated each discovery step, from training dataset creation, via graph-based generative design of optimized monomer units, to molecular dynamics simulation of gas permeation through the polymer membranes. For the latter, we have devised a representative elementary volume (REV) enabling permeability simulations at about 1000× the volume of an individual, ML-generated monomer, obtaining quantitative agreement. The discovery-to-validation time per polymer candidate is on the order of 100 h using one CPU and one GPU, offering a computational screening alternative prior to lab validation.
Similar content being viewed by others
Introduction
So far, the discovery of new materials has been a time consuming and resource intensive effort. The following trial-and-error approach is typically employed: identifying known materials with properties similar to the new material’s target properties and then modifying or combining them for achieving the desired outcome. The approach is driven by a specialist’s knowledge, laboratory experimentation, and it can take years to yield results. The computer revolution has brought about powerful simulation techniques, such as the density functional theory (DFT)1,2 method, that are aiding materials discovery today. High-throughput computational materials screening and design (HCMSD) methods have enabled substantial speed-up of the process3,4,5,6,7,8. However, one limitation of HCMSD is that it usually relies on time consuming ab initio calculations, such as DFT simulations4,5,7,8, for modeling the occurring physical and chemical processes. As a result, the large number of computations required for probing the phase space or performing materials screening can render HCMSD impractical.
The emergence of repositories with large sets of experimental and simulation data has enabled the application of ML methods as a data-driven pathway to materials discovery9,10,11,12. ML-based materials design10,13,14,15,16 has a potential advantage over HCMSD: while still relying on materials screening, it is not dependent solely on ab initio simulations of either classical or quantum mechanical molecular dynamics occurring in a chemical system. Recently, the inverse materials design (IMD) method17,18 has shown its potential: an algorithm creates optimized molecular structures based on a pre-defined feature vector containing a set of materials target properties. To complete the discovery process, the IMD output would have to undergo physical validation. For polymer membranes, this validation is needed at mesoscale where the process-relevant properties of amorphous materials emerge. At that scale, however, automated ab initio simulation methods for validating complex materials do not yet exist.
To exemplify the issue, let us consider the case of carbon dioxide separation in post-combustion applications. From a process perspective, polymer membranes19,20,21 have certain advantages, among them high tolerance for the challenging operating conditions and adaptability to the existing power plant steam cycle. However, a polymer’s gas filtration performance cannot be derived from the physical and chemical properties of the monomer constituents alone. Rather, it is determined by the heterogeneous internal structure and complex morphology of the amorphous polymer. Therefore, predicting and validating a membrane’s gas permeability remains a major challenge22.
Encouragingly, it was recently reported that machine learning applied to known polymer repeat units can predict gas separation performance of polymers that were not previously tested for these properties23. However, the reported method did not offer the IMD benefits of generating optimized monomer units and, therefore, could not generate new polymer candidates. Also, it lacked automated outcome validation of physical performance. In the following, we report a fully automatized, in silico materials discovery workflow that overcomes those limitations. For demonstrating the methodological advancements with regards to the discovery of small molecules, we have applied the workflow to the generative design and physical validation of polymers optimized for carbon dioxide filtration under realistic temperature and pressure conditions. We have limited our study to homo-polymers for which suitable experimental data and error margins are available. For copolymers, reliable experimental data on the spatial orientation and arrangement of the polymer chains are needed. Inclusion of molecular selectivity, such as carbon dioxide versus nitrogen, is currently limited by nitrogen force fields which overestimate the nitrogen uptake by at least 50%24, leading to unsatisfactory results. Novel force fields for nitrogen have been developed to address the issue in MOFs25, ZIFs26, and zeolites27; however, they are still lacking for polymers.
Results and discussion
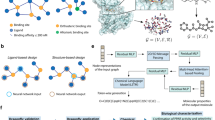
In Fig. 1, we show the discovery workflow end-to-end, from training dataset preparation, via ML generative design to physical validation by molecular dynamics simulation. Small organic molecules, or monomer units, that typically qualify as candidate building blocks for polymer membranes, are often treated as graphs and can be converted to computer readable SMILES format28. For training dataset preparation based on SMILES, we have extended a quantitative structure–property relationships approach29 and made it available through our polymer property prediction (PPP) engine. For ML generative modeling, we have created an IMD engine30 which extracts molecular features with regression and performs graph-based construction with SMILES input. Finally, the discovered monomers are physically validated at meso-scale by means of automated constant pressure difference molecular dynamics (CPDMD) simulations22, a non-equilibrium method suited for predicting a polymer membrane’s gas filtration performance under realistic process conditions. Overall, our workflow is consistent with the approach outlined in ref. 31.
The training dataset preparation sequence is shown in the left box of Fig. 1: polymer name collection from existing data sources, polymer name conversion into the OPSIN SMILES strings32, and polymer name mapping to suitable target polymer properties and their respective numerical values. As high-quality lab data is often sparse or not available at all, we have used PPP for calculating polymer properties based on topological variables, such as connectivity indices, combined with geometrical variables and other structural descriptors29. For polymer properties predicted with the PPP engine, the underlying QSPR regression models were trained on experimental data29. Therefore, there is no dependence of ML predictions on membrane thickness. In Fig. 2, we illustrate the PPP conception and outline as a representative example the prediction of the half-decomposition temperature Td,1/2 (see “Methods” section). In the example of poly(vinyl butyral) shown in Fig. 2b, we obtain Td,1/2 = 646 K which is in agreement with the experimental value of 645 K29. Similarly, we have used PPP to predict the glass transition temperature Tg (in K) and CO2 permeability (in Barrer) for all 1169 homo-polymers in our dataset shown in Fig. 2c. These are suitable target properties for informing generative design of new monomers to be validated in gas separation membranes at process level. Although the accuracy of property predictions by the PPP engine is limited in comparison with experimental data, see Supplementary Material and Supplementary Fig. 1, the output is useful for obtaining molecular discovery results.
a Example of half-decomposition temperature Td,1/2 which is calculated according to Eq. (1) (see “Methods” section). For Td,1/2, structure–property correlations were established with the first-order (bond) connectivity index 1χV, the number of hydrogen atoms NH and the number of vertices Nvertices in the hydrogen-suppressed graph representation of a polymer’s monomer29. b Example of a hydrogen-suppressed graph representation for poly(vinyl butyral) built from the polymer name and the corresponding OPSIN SMILES string32. (i) Schematic representation of poly(vinyl butyral); (ii) alternative representation with brackets not intersecting the bonds; (iii) hydrogen-suppressed version of (ii) with the valence connectivity indices δV in the vertices and (iv) with the bond indices βV in the edges, respectively (see Eqs. (2) and (3) in the “Methods” section). c Multi-dimensional property distribution of the input dataset containing 1169 homo-polymers.
The ML generative design sequence is shown in the middle box of Fig. 1: feature extraction and selection, regression model training, feature optimization, and graph-based structure generation. For automatically generating new monomers with pre-defined target properties, we have represented each of the homo-polymers in the input dataset by its monomer in the form of a feature vector. Each unique monomer is represented by one SMILES string and is encoded to a specific feature vector. As visualized in Fig. 3a, each feature vector contains structural descriptors such as the numbers of heavy atoms, rings, aromatic rings, substructures, and fingerprints.
a The structure of each monomer in the input dataset is encoded as a feature vector. Here, the aring feature label stands for aromatic ring and the numbers below each label indicate their respective occurrences. b Regression results for each of the pre-defined target properties: PCO2, Tg, and Td,1/2. Blue circles: training data, red line: data fit. c Feature vector representation with encoded molecular building blocks: atoms, rings, aromatic rings, sub-structures, and fingerprints. Decoding the feature vector reveals a pyridine molecule for which the SMILES representation is also shown. d Multi-dimensional property distribution of the generated dataset. e ML-generated monomers selected for physical validation in polymer representations via molecular dynamics simulation. The SMILES representation of each monomer is shown below the respective unit.
For molecular property prediction, we have trained and cross-validated regression models with respect to multiple sets of feature vectors and to each of the pre-defined target properties: half-decomposition temperature Td,1/2, glass transition temperature Tg, and CO2-permeability PCO2 as shown in Fig. 3b. The cross-validation (CV) process is performed by using the random data split method for training and testing purposes.
Specifically, we have trained six regression models: Lasso Regression, Ridge Regression, Elastic Net Regression, Random Forest Regression, Kernel Ridge Regression, and Support Vector Regression (SVR). For training each model, we have applied both hyperparameter optimization and feature selection. For the Kernel Ridge and SVR models, respectively, we have developed a new method that efficiently performs hyperparameter optimization and feature selection simultaneously (see the “Methods” section). For the other models, we have performed grid search for optimizing hyperparameters while selecting features using the SelectFromModel class in Scikit-learn33. In Supplementary Table 2, we show a statistics summary of CV scores for four regression models with respect to selected feature vector sets (see Supplementary Material for discussion). To maximize accuracy, we have selected the SVR model yielding the best crossvalidated R2 score. The parity plots in Fig. 3b represent the model with the best CV score, which is quantified in Supplementary Fig. 2. This demonstrates the effectiveness of both hyperparameter optimization and feature selection processes. The idea is to find newly created feature vectors which fit the capability of the optimized models. We show a baseline example with and without hyperparameter optimization in Supplementary Fig. 2, and the improvement of the R2 score is impressive. Note, that we have obtained both parity plots and CV scores from runs that were also used for hyperparameter optimization.
For generative design, we have then optimized the feature vectors through inversion of the prediction model within the pre-defined target property ranges which were set to: 550 K < Td,1/2 < 700 K; 400 K < Tg < 600 K and 630 barrer < PCO2 < 4000 barrer. We have expanded the optimized feature vectors to molecular structures through an advanced version of the Molecular-Customized McKay’s Canonical Construction Path Algorithm30,34,35. The algorithm repeats cycles of connecting structural fragments such as atoms, rings, and substructures, and cycles of feature screening. Our methodological advancements (see “Methods” section and Supplementary Information) enable the application of graph-based generative design to complex molecular structures. In addition, the IMD allows for defining design rules with regards to structural constraints, the range of the number of substructures, as well as fragment patterns. As a result, chemical subject matter expertise can inform the generative design process. In Fig. 3c, we have visualized an example of how the generative algorithm transforms a feature vector into a molecular structure. A feature vector encodes structure-specific information for each molecular building block. The algorithm generates a specific molecular expression of the feature vector based on a library of building blocks created during the feature vector encoding process. Note, that this decoding process is not bijective, and a feature vector can be decoded into various monomers.
After completing the ML-generative design sequence and screening our initial discovery results for target property range and discrepancies between predicted and calculated polymer property values, we have obtained a set of 784 new monomer candidates shown in Fig. 3d. We have suppressed duplication of the same monomer in the generation algorithm by applying the canonical construction path algorithm using a special assignment of head and tail atoms (based on * character in SMILES strings). As an improvement with regards to the initial data set, we demonstrate that about 50% of the generated monomers exhibit optimized properties that simultaneously fit the predefined target ranges. Specifically, in our input dataset, only two out of 1169 homo-polymers fulfill all requirements for Td,1/2, Tg, and \({P}_{C{O}_{2}}\). In the output data set, 390 generated polymers simultaneously fulfill the above requirements. The efficiency of the process, i.e., the number of newly generated homo-polymers that fulfill all target requirements divided by the total number of generated species, depends on how narrowly we define the range of the predefined target properties. As a rule-of-thumb, we can say that “the wider the target property range, the higher the generation efficiency.” In the present case, the overall generation process efficiency is around 50%. While 394 out of 784 newly generated species do not fulfill all requirements simultaneously, we find that those structures are nevertheless very close to the pre-defined target property ranges. Figure 3d shows a 2D plot with three parameters and the same data are shown in a 3D representation in Supplementary Fig. 5. For demonstrating the effectiveness of our inverse design approach, we show for comparison in Supplementary Fig. 6 the predicted properties of polymers generated through random recombination of building blocks in the training set by shuffling their feature vectors. In the following, we will physically validate the most promising of the discovered monomers, visualized in Fig. 3e, in a polymer membrane configuration by means of automated molecular dynamics simulation.
The automated molecular dynamics simulation sequence is shown in the right box of Fig. 1: creation of SMILES representation of the discovered monomer, creation of a polymer membrane representation with the discovered monomer, and physical simulation of the gas filtration process through the membrane. Prior to applying the above sequence to the newly generated monomers, we have confirmed the suitability of the CPDMD method for physical validation of membrane performance through extensive benchmark analyses with known polymers, (see “Methods” section and Supplementary Information). A fundamental question occurs with regards to the minimum volume that adequately represents the properties of complex materials at mesoscopic scales. In the present case of gas separation with polymer membranes, we have adopted the concept of representative elementary volume (REV), which is routinely used for characterizing porous media36. REV can be understood as the smallest material volume for which a physical property can be determined such that it yields a value that is representative of the bulk. To illustrate this concept, we show in Fig. 4a cross sections through computational membrane representations exhibiting porosity variations. Depending on the region sampled, a material’s porosity can be smaller or larger than the bulk average. If probed at or above REV level, the porosity value matches the bulk average.
a Cross-sectional view of a meso-scopic polymer membrane representation. The volumes rendered in green color are void spaces visualized using 3V56,57. The solid frame visualizes the representative elementary volume (REV) concept; the dashed frame does not adequately capture the membrane’s average porosity. b Representative monomers for determining the polymer REV. The asterisks indicate the head and tail atoms, respectively. c CO2 permeability of representative, ML-designed polymers. d CPDMD simulation of CO2 filtration dynamics. The shaded area indicates the transient regime.
To probe REV with regards to our polymer permeability simulations, we have investigated three representative polymers with relatively high permeability values: TDA1-DMN, PIM-PI-EA and IBPA shown in Fig. 4b—bottom of figure from left to right. For each of the three polymers, we have performed five independent CPDMD simulations using the simulation box set up shown in Fig. 1. By doubling the number of atoms in the simulation and keeping the membrane thickness fixed at 6nm, the cross-sectional area also doubled, from around 50 to 100 nm2. By probing larger areas and randomly sampling the amorphous polymeric chains making up the membrane, we observe a trend in Fig. 4b that the simulations with larger volume tend to approach the experimental values.
After establishing both the automated CPDMD simulation protocol and the REV determination (see “Methods” section and Supplementary Information), we have proceeded with the validation of the three shortlisted new polymer candidates generated by IMD. Two similar monomers were selected for investigating how head and tail positions influence the simulation results. For each of the three polymers, we have performed five independent CPDMD simulations using the simulation box in Fig. 1. As a key result of our investigation, we show in Fig. 4c the simulated CO2 permeability values obtained for the polymer membrane representations of ML-designed monomers. We observe quantitative agreement, within the error bars, of ML-predicted permeability values and the CPDMD-based physical validation results. To our knowledge, this is the first computational performance validation of an ML discovered, amorphous polymeric membrane material.
For analyzing the filtration dynamics, we show as a representative example for one of the polymers in Fig. 4d the simulated CO2 permeability along with the number of CO2 molecules permeated and sorbed, respectively, as function of simulation time. The corresponding CO2 density profile evolution is shown in Supplementary Fig. 13. Initially, the CO2 molecules are located at the left-hand side of the membrane, see simulation box in Fig. 1, which is connected to the gas feed chamber. As shown in Fig. 4d, CO2 molecules are penetrating the membrane at 1 ns. The membrane saturation level of roughly 100 CO2 molecules on average, corresponding to an average density of 0.06–0.07 g/cm3, is reached at about 5 ns. At 10 ns, the CO2 permeability has converged towards the saturation value of 5000 barrer. After 30 ns, the permeability fluctuations have disappeared and the membrane filtration has reached a steady state. The steady-state filtration regime is characterized by the constant slope of permeated CO2 molecules as function of time from which the permeability value can be extracted using Eq. (6) (see “Methods” section).
The main transport characteristics captured by the CPDMD simulations are (i) the interaction between gas molecules and polymer membrane which determines solubility and, consequently, the selectivity with regards to a specific gas molecule and (ii) the diffusion of gas molecules through the void spaces between packaged polymer chains which determines the membrane’s permeability. We note that while the thickness of manufactured polymer membranes are typically on the order of micrometers, the much thinner simulated membrane predicts experimental permeability values reasonably well, within the same order of magnitude. As expected, the accurate and repeatable determination of gas permeability is limited experimentally as well as theoretically and large error margins are an intrinsic characteristic associated with the properties of amorphous materials22.
Using a standard computational framework (one Intel Xeon E5-2667 CPU, one NVIDIA Tesla K80 GPU), the overall computation time, from dataset preparation to ML generation to physical validation, is of the order of 100 h for polymeric membranes with higher permeabilities (above 1000 barrer). A computational bottleneck currently exists for reaching gas saturation and steady state filtration in lower-permeability membranes as shown in Supplementary Fig. 12.
Future extensions of this work would benefit from advanced representations of a membrane’s morphology. One example would be to pack the monomers randomly in a virtual cubic box and connect their head and tail atoms according to predefined probabilities—instead of packing the polymer chains randomly. This would more closely resemble the actual polymer formation process. In generative modeling, the extension to molecular structures with higher complexity and the introduction of a user-defined objective function could open the pathway to the generation of polymers with higher complexity, such as block co-polymers. We expect that adding target properties for molecular selectivity to the optimization workflow and extending the generative algorithms to the design of co-polymers will further improve discovery outcomes.
In summary, we have reported fully automated, end-to-end computational discovery of polymer membranes for carbon dioxide separation. We have demonstrated each discovery step, from automated training data and feature vector creation via generative inverse design of new monomers to non-equilibrium molecular dynamics simulation of gas filtration by the polymer membrane. Molecular dynamics simulations successfully predict a polymer’s filtration dynamics and permeability if performed with a minimum representative volume of the complex material. For computationally designed polymers, we have obtained quantitative agreement between the CO2 permeability predictions by means of the ML models and the molecular dynamics-based, physical process simulations. Our work opens a pathway for advancing ML-generative design beyond small-molecule applications and will substantially accelerate the discovery of complex materials for scaled applications.
Methods
Polymer property calculation for automated training dataset generation
For creating the training dataset, we have collected representative homo-polymers names in IUPAC nomenclature standard, from multiple polymer classes as provided by the PolyInfo database37. We have then converted their individual monomer unit names to SMILES format (with their head and tail units tagged) using the Open Parser for Systematic IUPAC nomenclature (OPSIN)32. Based on our analysis of the gas separation process, we have selected three suitable figures-of-merits or target properties for polymer membranes: glass transition temperature (Tg in K), half-decomposition temperature (Td,1/2 in K), and permeability (P) for CO2 (in Barrer). Tg is the temperature above which segmental motions of polymer chains occur such that they negatively affect a polymer membrane’s mechanical stability. Tg also defines the transition limit between glassy and rubbery polymers (temperature below and above Tg, respectively). Glassy polymers dominate the Roberson upper bound38,39 due to higher solubility coefficient39, or, in other words, better selectivity. However, rubbery polymers have lower solubility and higher diffusion coefficients39, i.e higher permeability and lower selectivity. Similarly, Td,1/2 defined as the temperature at which the loss of weight during pyrolysis (at a constant rate of temperature rise) reaches 50% of its final value should be reasonably high as it is a measure for chemical stability. A high permeability for CO2 is desirable as a measure of the gas flux through the membrane. However, it is limited by a trade-off with the membrane’s selectivity \({P}_{C{O}_{2}}/{P}_{{N}_{2}}\). For creating the training dataset, we have calculated \({P}_{C{O}_{2}}\), Td,1/2, and Tg data using the PPP engine.
Calculation of Td,1/2:
Best structure–property correlations were established with first-order (bond) connectivity index 1χV; number of hydrogen atoms NH and number of vertices Nvertices in the hydrogen-suppressed graph representation of a polymer’s monomer29. The functional relation for Td,1/2 was obtained through a linear regression against the best correlation descriptors:
Figure 2b displays the calculation steps performed by the PPP engine for poly(vinyl butyral). Starting with the (i) hydrogen-suppressed graph representation of poly(vinyl butyral) monomer and its (ii) alternative representation with the square brackets not intersecting the bonds, the (iii) valence connectivity indices δV in the vertices and the (iv) bond indices βV in the edges are calculated according to Eqs. (2) and (3), respectively:
where ZV is the number of valence electrons of an atom, NH is the number of hydrogen atoms bonded to it, and Z is its atomic number. \({\beta }_{ij}^{V}\) is the product of δV at the two vertices (i and j), which define a given edge or bond.
The first-order (bond) connectivity index 1χV of the entire molecule is defined through the summation over the edges of the hydrogen-suppressed graph:
By combining Eqs. (1) and (4), counting the number of vertices and the hydrogen atoms and calculating the molar mass of poly(vinyl butyral), we obtain Td,1/2 = 646 K which is in agreement with the experimental value of 645 K29.
Hyperparameter optimization and limited discrepancy search
The procedure referred to as feature selection identifies a subset of features for achieving accurate predictions, rather than using the entire set of the original features40. In other words, feature selection allows a machine learning algorithm to learn a model in a lower-dimensional space. The dimensionality reduction typically leads to computational performance enhancements.
Hyperparameter optimization (HPO) is also key for enhancing the model performance. There are many HPO algorithms available in the literature, including grid search and Bayesian optimization, see for example, reference41. In theory, hyperparameter configurations are specific to a feature set used to train a machine learning model. One set of hyperparameter configurations that works well for one feature set might not be the best for another feature set. On the other hand, both feature selection and HPO typically require intensive computation. For example, given N features, finding an optimal feature requires (2N) possible feature sets. For M hyperparameters, each of which has b configurations after its possible values are discretized, there are (bM) possible choices for the hyperparameter configurations. An optimal feature set and hyperparameter configurations need to be found out of (2NbM) combinations. In practice, feature selection and HPO are performed separately to reduce the computational overhead, e.g., perform HPO after feature selection selects an optimized feature set with default hyperparameter configurations. However, this approach might not represent a good combination of the feature set and hyperparameter configurations.
For validating our model and tuning hyperparameters, we have optimized the average (R2) score of the threefold cross-validation (CV) with 10 repeats. To that end, we have developed a systematic local search algorithm that simultaneously performs feature selection and HPO for a non-linear machine learning model. This approach leads to an optimized hyperparameter configuration specific to a selected feature set. To reduce the computational overhead, our approach focuses only on small, promising search spaces where optimized solutions are likely to occur. We discretize possible values for each hyperparameter and formulate feature selection and HPO as a variable-value assignment task. This means that each variable corresponds to another variable to which one value needs to be assigned. The variable for a feature is set to either true or false, while the variable for each hyperparameter is set to one of the discretized hyperparameter-values.
Our approach is based on limited discrepancy search (LDS)42,43. The idea behind LDS has been studied in the artificial intelligence community and has a variety of applications such as in reference30,35. LDS starts with an initial solution, i.e., initial variable-value assignment, and keeps refining it until a satisfactory solution is obtained.
Our current implementation calculates the initial solution passed to LDS as follows: It first calculates optimized hyperparameter configurations based on grid search with the whole feature set. With these hyperparameter configurations, it then computes the initial feature set based on so-called sequential backward selection (SBS)40. Our SBS implementation starts with the whole feature set. It repeats a greedy elimination of one feature (without which a score is improved) until no further improvement is obtained.
The solution refinement step of LDS consists of a series of local search controlled by the notion of discrepancy. Given the current best solution bs, LDS assumes that a better solution exists in a search space whose solutions are similar to bs. In our implementation, the discrepancy for a solution s is defined as the number of variables whose assigned values have differences between bs and s. A smaller discrepancy indicates that s is more similar to bs.
LDS introduces a discrepancy threshold d and performs local search in an iterative manner. After setting bs to the initial solution calculated by SBS, LDS performs depth-first search with d=1 and attempts to find a better solution than bs in a search space where solutions are located that have a different value than bs only for one variable. If no better solution is found, LDS increments d and performs local search with d=2. If no better solution is found again, LDS performs search with d=3, and so on. If a better solution is found, LDS resets d=1 and bs to the better solution and restarts a local search with d=1. LDS repeats these steps until the allocated time is used up or d reaches a preset, maximum value.
There are several implementation choices for LDS to select a next variable for updating its value. Before performing a new iteration of local search, our current implementation orders variables in ascending order of the following formula: w1v(x) + w2u(x), where w1 and w2 are constants, v(x) is the number of times variable x is selected in local search, and u(x) is the number of times variable x fails to improve bs. This formula attempts to remain the values of the variables unchanged that have contributed to improving a score as well as to prioritize the variables that have not been explored sufficiently.For the purpose of this study, we have chosen w1=2 and w2=1.
In Supplementary Fig. 2, we show a comparison of regression results obtained with and without the application of hyperparameter optimization.
Because linear models and their variants tend to perform poorly in our application, we have tested an approach for performing feature selection (FS) + HPO based on LDS for obtaining a non-linear model with acceptable performance. As described in the Methods Section, the approach includes training and validation for each combination of feature sets and hyperparameters selected by LDS—without distinguishing the feature selection. Introducing nested CV to our LDS-based approach creates excessive computational costs. Therefore, we have chosen the approach presented in our manuscript.
For analyzing how data leakage affects model performance, we have performed a comparison between the original algorithm and an improved version. The results are summarized in Supplementary Table 3.
In summary, in our approach the regression models are designed to find relationships between feature vectors (FV) and target property values. Some of the holdout feature vectors will not be included during the training process. As a result, the prediction values for holdout data fit poorly. In Supplementary Fig. 3 and Supplementary Table 4, we show a comparison between regression models in which 25% of the dataset is randomly selected to be holdout data. Even if there is a model that fits both training and holdout data well, it does not necessarily lead to superior discovery results - as the FVs have hundreds of dimensions. For reducing the complexity of the generation process, it is more efficient to use models which include all the FVs of the dataset and then fine tune the molecular generation process.
Feature vector optimization
Based on graph theory and atomic configurations, there exist multiple feature types which can be combined for application of machine learning models, among them the number of heavy atoms, number of rings, substructures, fingerprints, Coulomb matrix, dipole moment, potential energy, and experimental conditions30.
We obtain suitable feature vectors by minimizing the distance to the target property value using a particle swarm optimization (PSO) algorithm, which optimizes a population of feature vectors starting from a set of randomly generated ones. By using Eq. (5), we estimate feature vector values fv based on a target property value vp and a regression model fp by minimizing the score of each feature vector v. More specifically, the minimization is performed over the square error of the estimated value which is normalized by the prediction variance \({\sigma }_{p}^{2}\) to which a penalty function is added to account for violations of structural constraints. The violation of structural constraints is evaluated by means of the realizability of a molecular structure connected by sub-structures in the corresponding feature vector. The evaluation of the violation of structural constraints is a required but not sufficient step for successfully decoding molecular structures. Although most feature vectors obtained by PSO may not be decoded, they are instructive for navigating the huge search space for the generative algorithm. The reason is that new feature vectors calculated by PSO may include sub-structures which differentiate them from the initial feature vector set. These new feature vectors might then enable the generation of new structures. The enhanced version of our generative algorithm allows for bypassing the PSO-assisted feature estimation. New molecular structures are evaluated by a regression function and will be kept as solutions if the estimated property values are within the pre-defined range.
Generative molecular design
The McKay’s Canonical Construction Path Algorithm generates molecular structures efficiently, exhaustively, and without isomorphic duplication. Starting from a single vertex, a molecular graph is augmented by adding a new vertex to extendable vertices of the graph. Generation of isomorphic molecular graphs is suppressed by pruning the search tree at the following steps in the generation:
-
1.
In generating a child search node, only one extendable vertex is chosen among a set of symmetric vertices in a molecular graph.
-
2.
After generating a child search node, the search node survives only when the generation step is a canonical augmentation.
We show in Supplementary Fig. 4a an example of a search tree for generating a molecular graph of 5 carbon atoms with single bonds from “CC(C)C.” The vertices are indexed by order of addition to the graph. A set of symmetric vertices are obtained as an orbit of automorphism of a graph by applying a canonical labeling algorithm. Orbits at the root node “CC(C)C” are 0 and 1, 2, 3, and vertices of 0 and 1 (a vertex of the smallest number in an orbit) are chosen for adding a new vertex (pruning 1). At the bottom of the search tree, isomorphic molecular graphs “CCC(C)(C)C” are generated, but only one of them should survive. A canonical labeling algorithm assigns labels of sequential numbers to vertices of a molecular graph, and “0” is assigned to one of the vertices of degree 1. We define that a graph augmentation is canonical if ‘0’ is assigned to the lastly added vertex by a canonical labeling algorithm. Therefore, the search node of an isomorphic molecular graph at the right branch is pruned in this example (pruning 2). When a search node survives, orbits of automorphism of the graph obtained from the canonical labeling algorithm can be used for choosing extendable vertices in the next generation step.
Since in our implementation of McKay’s generation algorithm only tree structures are generated with graph augmentation by adding an atom as a vertex, the graph augmentation is extended to use fragment structures, such as ring structures and user defined structures, as vertices to add44. A fragment structure is regarded as a huge atom, whose symbol is the SMILES of the structure, with valency in the graph augmentation. Only when vertices of a fragment structure are chosen for adding a new vertex, the fragment structure is expanded to the actual molecular graph. Since the canonical labeling is time consuming and its complexity depends on the size of a graph, treating fragment structures as vertices can improve the performance of the generation algorithm.
The advanced version of the generative algorithm35,44 inherits user-customized design constrains such as, for example, expected or unexpected sub-structures in SMILES format, and the inverse designed feature vectors such as, for example, the number of heavy atoms, rings, and occurrences of fragment structures, and then converts them into molecular structures. Constraint functions capture design rules such as, for example, disallowing triple bonds between carbon atoms, limiting the number of molecular rings in the structure to between 4 and 9, or including preferential molecular substructures. For the purpose of this study, all constraints have been merged with the extracted feature vectors and best regression models for subsequent iterations of optimized structure generation.
An example with a ring of six atoms is shown in Supplementary Fig. 4b. In a first step, the orbits of the automorphism group are obtained from the SMILES representation of a given sub-structure. We then create the isomorphic equivalent graph by replacing the atom name with the SMILES name and the minimum index number (indices 1 and 3). In this step, those vertices without “free hand” are eliminated which helps identifying the symmetry of the graph. For better handling, the isomorphic equivalent graph is further simplified to a single vertex representation by selecting vertices with minimum index number in each orbit, whereas other vertices are replaced by dummy atoms. Finally, we obtain the orbits of the automorphism group and the minimum index number of each orbit is selected to be an extending vertex of the sub-structure.
Supplementary Fig. 4c shows a construction example. During the generation of a molecular structure as a colored graph (graph of various atoms) and by adding a vertex with a connecting edge one by one, the algorithm minimizes the number of vertices in order to improve the performance of the canonical labeling which is a bottleneck routine of the process. In the root graph, an extending vertex which has a minimum label in an orbit of an automorphism is considered to be a single vertex graph. In order to extend the vertices, it is replaced by an isomorphic equivalent representation. The new vertex is extended and canonical labeling of the entire graph is performed. Once the canonical construction path is validated, the original representation will be recovered. The new structure will be tested against the pre-defined design constraints. The cycle repeats until it fulfills pre-set requirements such as number of generated results with pre-defined target property values.
Note, that the generative model (GM) algorithm we have developed is based on training feature vectors (FVs) against target property values. Some advantages in comparison to widely used DNN (deep neural network) based methodologies are discussed in ref. 30. In short, DNN based GMs require large amounts of data and long training times, but they are capable of efficiently generating large quantities of new candidate molecules from a trained hyperspace.
In FV-based GMs, the training time is typically much shorter than in DNN based GMs. However, the generation process has more flexibility and uncertainty. For example, if the generation results lack variety, we could increase the depth of search tree and the beam search size. In return, this choice would make the generation time harder to predict as single molecules or sub-structures are randomly selected from inversely calculated FVs. We conclude that a straightforward comparison between the two concepts (FV-based versus DNN-based GMs) is complicated and the best suited approach should be chosen in view of a specific application. Particularly in cases of data scarcity, we believe the FV-based method provides a good balance between model precision and generative design flexibility.
Computational representation of the polymer membrane
For the physical validation of ML predicted CO2 permeability, we have created a method to automatically design a polymer membrane representation which is suitable for molecular dynamics simulation, see right box in Fig. 1. In a first step, the SMILES strings of ML designed monomers are indexed to indicate the position of head and tail atoms so they can be used as input for PySIMM45,46. We have then used the Force Field Assisted Linear Self-Avoiding Random Walk application in PySIMM45 to build a linear polymer chain with a maximum number of about 800 heavy atoms which are defined as atoms other than hydrogen. This way, we have kept the length of the polymer chain rather constant, independent of the monomer size. For describing the interactions between intra-chain and inter-chain atoms, we have used the DREIDING force field47.
Once the chain building step is completed, PySIMM saves the LAMMPS48 topology file with the associated force field parameters. Then, the polymer chains are packaged in a 3D box using Packmol49. The 3D simulation box is periodic in x, y, z directions. We are aware of the limited accuracy of applying force-field parameters generated automatically by PySIMM for polymer modeling, and opls-aa parameters50 can be adopted for an improved accuracy. For defining the membrane thickness, the z dimension of the box is set to 6 nm (see Supplementary Material for more details). The dimensions of the box in x and y are defined by a multiplication factor of the polymer chain size. The number of polymer chains is defined by the total number of atoms in the polymer membrane—20,000 in the present case. To keep the membrane thickness in z-direction fixed at 6nm, rigid walls are placed in the x, y membrane planes. To avoid interactions between periodic images in z-direction, a vacuum layer with a thickness of 5 nm is placed at each side of the polymer membrane.
The system then undergoes an equilibration process that consists of a nine-step compression-relaxation sequence, similar to the approach in ref. 22: (1) energy minimization with isothermal and isochoric (NVT) MD simulation at 1 K for 100 ps, (2) NVT MD simulation at 300 K for 100 ps, (3) isothermal and isobaric (NPT) MD simulation at 300 K and 1 atm for 100 ps, (4) NPT MD simulation at 300 K and from 1 atm to 3000 atm for 100 ps, (5) NPT MD simulation at 300 K and 3000 atm for 300 ps, (6) NVT MD simulation at 800 K for 100 ps, (7) NVT MD simulation at 300 K for 100 ps, (8) NPT MD simulation at 300 K and 1000 atm for 300 ps, the steps (6)-(8) repeats 30 times, and (9) NPT MD simulation at 300 K and 1 atm for 10,000 ps.
To account for long-range electrostatic interactions, we have adopted the reciprocal space Particle-Particle Particle-Mesh (PPPM) method. For all calculations, we have used 1 fs time steps and a cutoff radius of 1.4 nm for van der Waals and Coulomb interactions, respectively. To control temperature and pressure, we have used Nose–Hoover thermostats and barostats with a relaxation time of 0.1 and 1 ps, respectively.
All MD simulations were carried out with the LAMMPS package48,51,52,53. For further information regarding the effects of chosen force fields, chain lengths, membrane thicknesses and the equilibration process protocol, see Supplementary Information and Supplementary Figs. 8–10.
Automated membrane validation with molecular dynamics simulation
Two types of molecular dynamics (MD) simulations methods have been used to investigate transport through membranes: equilibrium MD (EMD) and non-equilibrium MD (NEMD). NEMD is ideally suited to represent an experimental membrane system in which an external driving force, such as a chemical potential or pressure gradient, is applied to the membrane. Specifically, we have chosen CPDMD to evaluate membrane based gas filtration22.
For benchmarking purpose, as shown in Supplementary Fig. 7, we have chosen representative homo-polymers covering a broad CO2 permeability range. For six of these homo-polymers, we have performed five independent CPDMD simulations each using the simulation box set up in Fig. 1. The results are shown in Supplementary Fig. 11. Overall, we obtain reasonable agreement with literature values for BZ-CF3, IBPA, PIM-PI-EA and PEO, despite the large error bars for BZ-CF3 and PEO. The simulated CO2 permeabilities of TDA1-DM and PI-5 are higher than the literature values, however, one of the PI-5 samples is close to the experimental value. We note that due to the amorphous nature of polymers, both experimental and simulations results typically exhibit large error bars22.
To set up a CPDMD simulation, we have placed the membrane at the center of the simulation box with a fixed, rigid wall at each side of the membrane, 10 nm away from its surface, as shown in Fig. 1. To avoid interactions with periodic images in z-direction, we have placed a 5 nm vacuum layer beyond each rigid wall. The carbon atoms in the 5 Å surface layer of the membrane were fixed in z-direction by a harmonic potential with a force constant of 5.0 Kcal/mol Å2. Following ref. 22, we have estimated the number of CO2 molecules in the feed chamber using the ideal gas law NCO2 = NApV/RT, where NA is the Avogadro’s constant, R is the gas constant, p is the pressure set to 10 atm, T is the temperature set to 300 K, and V is the feed chamber volume, see Fig. 1. We have then performed NVT MD simulations at 300 K. Due to the pressure gradient, CO2 molecules are absorbed within the membrane and, subsequently, transported to the permeate side. To maintain the same initial pressure gradient of 10 atm, we have added CO2 molecules into the feed chamber while removing the molecules at the permeate side to produce a pseudo vacuum. We have run the addition/removal processes in cycles with a time interval of 200 ps following ref. 54. We have used the DREIDING force field47 for describing the interactions between intra-chains and inter-chains atoms. For CO2 molecules, we have used the rigid model TraPPE force field55. For the CO2/polymer LJ interactions, we have applied the Lorentz–Berthelot mixing rules. All MD simulations were performed with the LAMMPS package48,51,52,53 using the same parameters described in the previous “Methods” subsection.
From the NCO2 − t slope, the permeability PCO2 can be estimated following
where ΔNCO2 is the number of CO2 molecules permeated within time duration Δt, NA is Avogadro’s constant, l and A are the membrane thickness and area, respectively, and p is the partial pressure—10 atm in this case—in the feed chamber.
The termination criterion for CPDMD simulations is discussed in detail in the Supplementary Information and shown in Supplementary Fig. 12. The evolution of the simulated CO2 density profile across a polymer membrane is shown in Supplementary Fig. 13, complementing the simulation results shown in Fig. 4d for the same polymer.
Data availability
The training dataset containing polymer candidates in SMILES format is available—under https://doi.org/10.24435/materialscloud:64-c8—at https://archive.materialscloud.org/record/2023.20. The input data for calculating PCO2; Td,1/2 and Tg with the PPP engine can be found in the second and third column, respectively, and the output data in the remaining columns of the file PCO2-Tg-Thd-data-all-simulated.csv. This file is available—under https://doi.org/10.24435/materialscloud:64-c8—at https://archive.materialscloud.org/record/2023.20. The dataset containing AI discovered polymer candidates in SMILES format is available—under https://doi.org/10.24435/materialscloud:64-c8—at https://archive.materialscloud.org/record/2023.20.
Code availability
The Polymer Property Prediction (PPP) Engine is available at https://github.com/IBM/polymer_property_prediction. The jupyter notebook for polymer property predictions based on SMILES input is available—under https://doi.org/10.24435/materialscloud:64-c8—at https://archive.materialscloud.org/record/2023.20. The open-source version of Inverse Materials Design engine is available at https://github.com/GT4SD/molgx-core/. The jupyter notebook for the training and regression models with the open-source version of IMD is available—under https://doi.org/10.24435/materialscloud:64-c8—at https://archive.materialscloud.org/record/2023.20.
References
Hohenberg, P. & Kohn, W. Inhomogeneous electron gas. Phys. Rev. 136, B864–B871 (1964).
Kohn, W. & Sham, L. J. Self-consistent equations including exchange and correlation effects. Phys. Rev. 140, A1133–A1138 (1965).
Zhang, L., d’Avezac, M., Luo, J.-W. & Zunger, A. Genomic design of strong direct-gap optical transition in Si/Ge core/multishell nanowires. Nano Lett. 12, 984–991 (2012).
Jain, A. et al. A high-throughput infrastructure for density functional theory calculations. Comput. Mater. Sci. 50, 2295–2310 (2011).
Zhang, W., Sun, P. & Sun, S. A precise theoretical method for high-throughput screening of novel organic electrode materials for Li-ion batteries. J. Materiomics 3, 184–190 (2017).
Mounet, N. et al. Two-dimensional materials from high-throughput computational exfoliation of experimentally known compounds. Nat. Nanotechnol. 13, 246–252 (2018).
Horton, M. K., Montoya, J. H., Liu, M. & Persson, K. A. High-throughput prediction of the ground-state collinear magnetic order of inorganic materials using density functional theory. npj Comput. Mater. https://doi.org/10.1038/s41524-019-0199-7 (2019).
Brunin, G., Ricci, F., Ha, V.-A., Rignanese, G.-M. & Hautier, G. Transparent conducting materials discovery using high-throughput computing. npj Comput. Mater. https://doi.org/10.1038/s41524-019-0200-5 (2019).
Curtarolo, S. et al. The high-throughput highway to computational materials design. Nat. Mater. 12, 191–201 (2013).
Ramprasad, R., Batra, R., Pilania, G., Mannodi-Kanakkithodi, A. & Kim, C. Machine learning in materials informatics: recent applications and prospects. npj Comput. Mater. https://doi.org/10.1038/s41524-017-0056-5 (2017).
Hafiz, H. et al. A high-throughput data analysis and materials discovery tool for strongly correlated materials. npj Comput. Mater. https://doi.org/10.1038/s41524-018-0120-9 (2018).
Cai, J., Chu, X., Xu, K., Li, H. & Wei, J. Machine learning-driven new material discovery. Nanoscale Adv. 2, 3115–3130 (2020).
Liu, Y., Zhao, T., Ju, W. & Shi, S. Materials discovery and design using machine learning. J. Materiomics 3, 159–177 (2017).
Lu, W., Xiao, R., Yang, J., Li, H. & Zhang, W. Data mining-aided materials discovery and optimization. J. Materiomics 3, 191–201 (2017).
Mannodi-Kanakkithodi, A. et al. Scoping the polymer genome: a roadmap for rational polymer dielectrics design and beyond. Mater. Today 21, 785–796 (2018).
Kim, C., Chandrasekaran, A., Huan, T. D., Das, D. & Ramprasad, R. Polymer genome: a data-powered polymer informatics platform for property predictions. J. Phys. Chem. C 122, 17575–17585 (2018).
Kim, K. et al. Deep-learning-based inverse design model for intelligent discovery of organic molecules. npj Comput. Mater. https://doi.org/10.1038/s41524-018-0128-1 (2018).
Wu, S. et al. Machine-learning-assisted discovery of polymers with high thermal conductivity using a molecular design algorithm. npj Comput. Mater. https://doi.org/10.1038/s41524-019-0203-2 (2019).
ichiro Noro, S. et al. Porous coordination polymers with ubiquitous and biocompatible metals and a neutral bridging ligand. Nat. Commun. https://doi.org/10.1038/ncomms6851 (2015).
Firpo, G. et al. The role of surfaces in gas transport through polymer membranes. Polymers 11, 910 (2019).
Powell, C. E. & Qiao, G. G. Polymeric CO2/N2 gas separation membranes for the capture of carbon dioxide from power plant flue gases. J. Membr. Sci. 279, 1–49 (2006).
Kong, X. & Liu, J. An atomistic simulation study on POC/PIM mixed-matrix membranes for gas separation. J. Phys. Chem. C 123, 15113–15121 (2019).
Barnett, J. W. et al. Designing exceptional gas-separation polymer membranes using machine learning. Sci. Adv. 6, eaaz4301 (2020).
Provost, B. An Improved N2 Model for Predicting Gas Adsorption in MOFs and Using Molecular Simulation to Aid in the Interpretation of SSNMR Spectra of MOFs. Master’s thesis, Université d’Ottawa/University of Ottawa (2015).
Dzubak, A. L. et al. Ab initio carbon capture in open-site metal–organic frameworks. Nat. Chem. 4, 810–816 (2012).
McDaniel, J. G. & Schmidt, J. R. Robust, transferable, and physically motivated force fields for gas adsorption in functionalized zeolitic imidazolate frameworks. J. Phys. Chem. C 116, 14031–14039 (2012).
Wang, S., Hou, K. & Heinz, H. Accurate and compatible force fields for molecular oxygen, nitrogen, and hydrogen to simulate gases, electrolytes, and heterogeneous interfaces. J. Chem. Theory Comput. 17, 5198–5213 (2021).
Weininger, D. Smiles, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 31–36 (1988).
Bicerano, J. Prediction of Polymer Properties - Third Edition (Marcel Dekker Inc., 2002).
Takeda, S. et al. Molecular inverse-design platform for material industries. In Proc. 26th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (ACM, 2020).
Pyzer-Knapp, E. O. et al. Accelerating materials discovery using artificial intelligence, high performance computing and robotics. npj Comput. Mater. https://doi.org/10.1038/s41524-022-00765-z (2022).
OPSIN. Open parser for systematic IUPAC nomenclature. https://opsin.ch.cam.ac.uk (2021).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Takeda, S. et al. AI-driven inverse design system for organic molecules. Preprint at arXiv https://arxiv.org/abs/2001.09038 (2020).
Takeda, S. et al. Molecule generation experience: an open platform of material design for public users. Preprint at arXiv https://arxiv.org/abs/2108.03044 (2021).
Costanza-Robinson, M. S., Estabrook, B. D. & Fouhey, D. F. Representative elementary volume estimation for porosity, moisture saturation, and air-water interfacial areas in unsaturated porous media: data quality implications. Water Resourc. Res. https://doi.org/10.1029/2010wr009655 (2011).
Polymer Database (PoLyInfo). https://polymer.nims.go.jp/en/ (2020).
Robeson, L. M. The upper bound revisited. J. Membr. Sci. 320, 390–400 (2008).
Robeson, L. M., Liu, Q., Freeman, B. D. & Paul, D. R. Comparison of transport properties of rubbery and glassy polymers and the relevance to the upper bound relationship. J. Membr. Sci. 476, 421–431 (2015).
Chandrashekar, G. & Sahin, F. A survey on feature selection methods. Comput. Electric. Eng. 40, 16–28 (2014).
Yang, L. & Shami, A. On hyperparameter optimization of machine learning algorithms: theory and practice. Neurocomputing 415, 295–316 (2020).
Harvey, W. D. & Ginsberg, M. L. Limited discrepancy search. In Proc. 14th International Joint Conference on Artificial Intelligence (IJCAI), Vol. 1. 607–615 (IJCAI, 1995).
Korf, R. E. Improved limited discrepancy search. In Proc. 13th National Conference on Artificial Intelligence (AAAI), Vol. 1. 286–291 (AAAI, 1996).
Hama, T. Molecular struture generation with substructure representations. U.S. Patent Application Publication US2020/0226804A1 (2020).
Fortunato, M. E. & Colina, C. M. pysimm: A python package for simulation of molecular systems. SoftwareX 6, 7–12 (2017).
Fortunato, M. E. & Colina, C. M. Pysimm. https://github.com/polysimtools/pysimm (2021).
Mayo, S. L., Olafson, B. D. & Goddard, W. A. DREIDING: a generic force field for molecular simulations. J. Phys. Chem. 94, 8897–8909 (1990).
Plimpton, S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 117, 1–19 (1995).
Martínez, L., Andrade, R., Birgin, E. G. & Martínez, J. M. PACKMOL: a package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 30, 2157–2164 (2009).
Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996).
Brown, W. M., Wang, P., Plimpton, S. J. & Tharrington, A. N. Implementing molecular dynamics on hybrid high performance computers - short range forces. Comp. Phys. Commun. 182, 898–911 (2011).
Brown, W. M., Kohlmeyer, A., Plimpton, S. J. & Tharrington, A. N. Implementing molecular dynamics on hybrid high performance computers - particle-particle particle-mesh. Comp. Phys. Commun. 183, 449–459 (2012).
W. M. Brown, Y. M. Implementing molecular dynamics on hybrid high performance computers: three-body potentials. Comp. Phys. Commun. 184, 2785–2793 (2013).
Liu, J. & Jiang, J. Molecular design of microporous polymer membranes for the upgrading of natural gas. J. Phys. Chem. C 123, 6607–6615 (2019).
Potoff, J. J. & Siepmann, J. I. Vapor–liquid equilibria of mixtures containing alkanes, carbon dioxide, and nitrogen. AIChE J. 47, 1676–1682 (2001).
Voss, N. R. & Gerstein, M. 3v: Cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555–W562 (2010).
3V: Voss Volume Voxelation. http://3vee.molmovdb.org/ (2020).
Acknowledgements
We acknowledge discussion with and support by Manuela F. B. Rodriguez, Rong Chang, Daiju Nakano, and Bruno Flach (all IBM Research).
Author information
Authors and Affiliations
Contributions
R.G. co-developed the PPP engine, developed the computational process of physical validation with molecular dynamics simulations, provided figures, and co-wrote the paper. H.H. developed the computational process from dataset preparation to ML Generative Design, performed ML Generative Design, and provided figures. A.K. developed algorithms for molecular design and co-wrote the paper. T.H. developed the advanced version of the generative molecular design algorithm. R.F.N. co-developed and open-sourced the PPP engine. B.L. improved the automatic force field parameter assignment. S.T. co-wrote the paper. L.H. performed data analysis. M.B.S. conceived and co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Giro, R., Hsu, H., Kishimoto, A. et al. AI powered, automated discovery of polymer membranes for carbon capture. npj Comput Mater 9, 133 (2023). https://doi.org/10.1038/s41524-023-01088-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-023-01088-3
This article is cited by
-
Deep reinforcement learning for microstructural optimisation of silica aerogels
Scientific Reports (2024)
-
Exploring the role of computer vision in product design and development: a comprehensive review
International Journal on Interactive Design and Manufacturing (IJIDeM) (2024)