Abstract
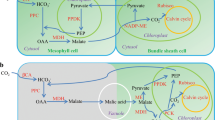
In C4 grasses of agronomical interest, malate shuttled into the bundle sheath cells is decarboxylated mainly by nicotinamide adenine dinucleotide phosphate (NADP)-malic enzyme (C4-NADP-ME). The activity of C4-NADP-ME was optimized by natural selection to efficiently deliver CO2 to Rubisco. During its evolution from a plastidic non-photosynthetic NADP-ME, C4-NADP-ME acquired increased catalytic efficiency, tetrameric structure and pH-dependent inhibition by its substrate malate. Here, we identified specific amino acids important for these C4 adaptions based on strict differential conservation of amino acids, combined with solving the crystal structures of maize and sorghum C4-NADP-ME. Site-directed mutagenesis and structural analyses show that Q503, L544 and E339 are involved in catalytic efficiency; E339 confers pH-dependent regulation by malate, F140 is critical for the stabilization of the oligomeric structure and the N-terminal region is involved in tetramerization. Together, the identified molecular adaptations form the basis for the efficient catalysis and regulation of one of the central biochemical steps in C4 metabolism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Amthor, J. S. From sunlight to phytomass: on the potential efficiency of converting solar radiation to phyto-energy. New Phytol. 188, 939–959 (2010).
Osborne, C. P. & Sack, L. Evolution of C4 plants: a new hypothesis for an interaction of CO2 and water relations mediated by plant hydraulics. Philos. Trans. R. Soc. Lond. B 367, 583–600 (2012).
Sage, R. F., Sage, T. L. & Kocacinar, F. Photorespiration and the evolution of C-4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47 (2012).
Sage, R. F., Christin, P. A. & Edwards, E. J. The C(4) plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169 (2011).
Emms, D. M., Covshoff, S., Hibberd, J. M. & Kelly, S. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol. Biol. Evol. 33, 1796–1806 (2016).
Maier, A., Zell, M. B. & Maurino, V. G. Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C(4) and C(3) photosynthesis. J. Exp. Bot. 62, 3061–3069 (2011).
Maurino, V. G., Saigo, M., Andreo, C. S. & Drincovich, M. F. Non-photosynthetic ‘malic enzyme’ from maize: a constituvely expressed enzyme that responds to plant defence inducers. Plant Mol. Biol. 45, 409–420 (2001).
Saigo, M. et al. Maize recombinant non-C4 NADP-malic enzyme: a novel dimeric malic enzyme with high specific activity. Plant Mol. Biol. 55, 97–107 (2004).
Tausta, S. L., Coyle, H. M., Rothermel, B., Stiefel, V. & Nelson, T. Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol. Biol. 50, 635–652 (2002).
Christin, P. A., Samaritani, E., Petitpierre, B., Salamin, N. & Besnard, G. Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol. Evol. 1, 221–230 (2009).
Saigo, M., Alvarez, C. E., Andreo, C. S. & Drincovich, M. F. Plastidial NADP-malic enzymes from grasses: unraveling the way to the C4 specific isoforms. Plant Physiol. Biochem. 63, 39–48 (2013).
Ashton, A. R. NADP-malic enzyme from the C4 plant Flaveria bidentis: nucleotide substrate specificity. Arch. Biochem. Biophys. 345, 251–258 (1997).
Iglesias, A. A. & Andreo, C. S. Kinetic and structural properties of NADP-malic enzyme from sugarcane leaves. Plant Physiol. 92, 66–72 (1990).
Detarsio, E., Wheeler, M. C., Campos Bermudez, V. A., Andreo, C. S. & Drincovich, M. F. Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at the putative nucleoside-binding sites. J. Biol. Chem. 278, 13757–13764 (2003).
Detarsio, E., Alvarez, C. E., Saigo, M., Andreo, C. S. & Drincovich, M. F. Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme. J. Biol. Chem. 282, 6053–6060 (2007).
Werdan, K., Heldt, H. W. & Milovancev, M. The role of pH in the regulation of carbon fixation in the chloroplast stroma. Studies on CO2 fixation in the light and dark. Biochim. Biophys. Acta 396, 276–292 (1975).
Fahnenstich, H. et al. Alteration of organic acid metabolism in Arabidopsis overexpressing the maize C(4)NADP-malic enzyme causes accelerated senescence during extended darkness. Plant Physiol. 145, 640–652 (2007).
Zell, M. B. et al. Analysis of Arabidopsis with highly reduced levels of malate and fumarate sheds light on the role of these organic acids as storage carbon molecules. Plant Physiol. 152, 1251–1262 (2010).
Matsumura, H. et al. Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure 10, 1721–1730 (2002).
Blasing, O. E., Westhoff, P. & Svensson, P. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275, 27917–27923 (2000).
Engelmann, S., Blasing, O. E., Westhoff, P. & Svensson, P. Serine 774 and amino acids 296 to 437 comprise the major C4 determinants of the C4 phosphoenolpyruvate carboxylase of Flaveria trinervia. FEBS Lett. 524, 11–14 (2002).
Jacobs, B., Engelmann, S., Westhoff, P. & Gowik, U. Evolution of C(4) phosphoenolpyruvate carboxylase in Flaveria: determinants for high tolerance towards the inhibitor l-malate. Plant Cell Environ. 31, 793–803 (2008).
Paulus, J. K., Schlieper, D. & Groth, G. Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4, 1518 (2013).
Xu, Y., Bhargava, G., Wu, H., Loeber, G. & Tong, L. Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases. Structure 7, 877–889 (1999).
Yang, Z., Floyd, D. L., Loeber, G. & Tong, L. Structure of a closed form of human malic enzyme and implications for catalytic mechanism. Nat. Struct. Biol. 7, 251–257 (2000).
Yang, Z. et al. Potent and competitive inhibition of malic enzymes by lanthanide ions. Biochem. Biophys. Res. Commun. 274, 440–444 (2000).
Yang, Z., Lanks, C. W. & Tong, L. Molecular mechanism for the regulation of human mitochondrial NAD(P)+- dependent malic enzyme by ATP and fumarate. Structure 10, 951–960 (2002).
Tao, X., Yang, Z. & Tong, L. Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism. Structure 11, 1141–1150 (2003).
Yang, Z. et al. Structural studies of the pigeon cytosolic NADP(+)-dependent malic enzyme. Protein Sci. 11, 332–341 (2002).
Alvarez, C. E. et al. The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme. Acta Crystallogr. D 74, 332–340 (2018).
Chang, G. G. & Tong, L. Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 42, 12721–12733 (2003).
Roalson, E. H. in C4 Photosynthesis and Related CO 2 Concentrating Mechanisms (eds Raghavendra, A. S. & Sage, R. F.) 319–338 (Kluwer, 2011).
Hsieh, J. Y., Chen, S. H. & Hung, H. C. Functional roles of the tetramer organization of malic enzyme. J. Biol. Chem. 284, 18096–18105 (2009).
Izui, K., Matsumura, H., Furumoto, T. & Kai, Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu. Rev. Plant Biol. 55, 69–84 (2004).
Maurino, V. G. & Weber, A. P. Engineering photosynthesis in plants and synthetic microorganisms. J. Exp. Bot. 64, 743–751 (2013).
Covshoff, S. & Hibberd, J. M. Integrating C4 photosynthesis into C3 crops to increase yield potential. Curr. Opin. Biotechnol. 23, 209–214 (2012).
Hibberd, J. M., Sheehy, J. E. & Langdale, J. A. Using C-4 photosynthesis to increase the yield of rice –rationale and feasibility. Curr. Opin. Plant Biol. 11, 228–231 (2008).
Kajala, K. et al. Strategies for engineering a two-celled C(4) photosynthetic pathway into rice. J. Exp. Bot. 62, 3001–3010 (2011).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Loytynoja, A. & Goldman, N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA 102, 10557–10562 (2005).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
Bricogne, G. et al. BUSTER v.2.9 (Global Phasing Ltd, 2009).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
van Stokkum, I. H., Spoelder, H. J., Bloemendal, M., van Grondelle, R. & Groen, F. C. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 191, 110–118 (1990).
Provencher, S. W. & Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 (1981).
Sreerama, N. & Woody, R. W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 (2000).
Whitmore, L. & Wallace, B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 (2004).
Schuck, P. & Rossmanith, P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers 54, 328–341 (2000).
Brautigam, C. A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015).
Acknowledgements
This work was funded by grants of the European Union (3to4) to V.G.M. and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany´s Excellence Strategy (EXC 2048/1, Project ID: 390686111 and EXC 1028, to V.G.M. and M.J.L. We acknowledge the European Synchrotron Radiation Facility (ESRF) for provision of synchrotron radiation facilities and we wish to thank L. Gordon for assistance in using beamline ID23-1. The Center for Structural Biology of Mercosur granted funding to C.E.A. for data collection at Institut Pasteur de Montevideo (IPM). We are grateful to N. Larrieux at the Protein Crystallography Facility IPM for assistance with crystallization and data collection.
Author information
Authors and Affiliations
Contributions
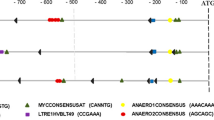
V.G.M. conceived and led the project and, together with M.F.D., designed and supervised the work and analysed data. C.-C.W., A.Bo. and M.S. produced the ZmC4- and ZmnonC4-NADP-ME recombinant proteins and obtained kinetic and structural data. A.H. planned crystallization studies of ZmC4-NADP-ME, collected X-ray diffraction data and solved, refined and analysed the structure. A.Bo. and A.H. produced ZmC4-NADP-ME crystals. C.E.A., F.T. and A.Bu. crystallized SbC4-NADP-ME, collected X-ray diffraction data and solved, refined and analysed the structure. C.-C.W. and M.J.L. designed the algorithm to identify strictly differentially conserved amino acid residues and performed the bioinformatic analyses. T.Z. and L.N.-S. performed and analysed circular dichroism and analytical ultracentrifugation analysis. All authors contributed equally to writing the manuscript and generation of the Figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nature Plants thanks Robert Furbank and Liang Tong and other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussion, Supplementary Figs. 1–8, Supplementary Tables 1–4, Supplementary Video legends.
Supplementary Video 1
3D overview of the overall SbC4-NADP-ME structure and the position of the mutated amino acids. The first section (1´´–30´´) shows the overall structure with the active site in each monomer, and the tilted position of each dimer in the dimer–dimer quaternary structure. The second section (30´´–60´´) shows the 3D position of the four mutated amino acids (F140, E339, Q503 and L544).
Supplementary Video 2
3D view of the interface connection between monomers in SbC4-NADP-ME. The first section (1´´–30´´) shows the position of F140 in monomers A and B. The second section (30´´-60´´) shows the position of the N-terminal region in the contact interface between each monomer. The N termini were selected such that they range from amino acid 84, the initial residue obtained in the SbC4-NADP-ME crystal structure, to amino acid 102 (corresponding to the chimeric proteins produced in ref. 15). Only selected residues/moieties are shown for clarity.
Supplementary Video 3
3D view of putative malate allosteric binding site (0´´–40´´). Shown are different views of the region surrounding E339 with its possible points of connection with active sites in SbC4-NADP-ME and the open/close switch conformation that is essential for catalysis.
Rights and permissions
About this article
Cite this article
Alvarez, C.E., Bovdilova, A., Höppner, A. et al. Molecular adaptations of NADP-malic enzyme for its function in C4 photosynthesis in grasses. Nat. Plants 5, 755–765 (2019). https://doi.org/10.1038/s41477-019-0451-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-019-0451-7
This article is cited by
-
Enhancing soil health and carbon sequestration through phytogenic treatment: insights into microbial functional pathways in pasture dieback affected soil
Plant and Soil (2024)
-
Role of C4 photosynthetic enzyme isoforms in C3 plants and their potential applications in improving agronomic traits in crops
Photosynthesis Research (2022)
-
Structural insights into the allosteric site of Arabidopsis NADP-malic enzyme 2: role of the second sphere residues in the regulatory signal transmission
Plant Molecular Biology (2021)