Abstract
The R-type voltage-gated Ca2+ (Cav) channels Cav2.3, widely expressed in neuronal and neuroendocrine cells, represent potential drug targets for pain, seizures, epilepsy, and Parkinson’s disease. Despite their physiological importance, there have lacked selective small-molecule inhibitors targeting these channels. High-resolution structures may aid rational drug design. Here, we report the cryo-EM structure of human Cav2.3 in complex with α2δ−1 and β3 subunits at an overall resolution of 3.1 Å. The structure is nearly identical to that of Cav2.2, with VSDII in the down state and the other three VSDs up. A phosphatidylinositol 4,5-bisphosphate (PIP2) molecule binds to the interface of VSDII and the tightly closed pore domain. We also determined the cryo-EM structure of a Cav2.3 mutant in which a Cav2-unique cytosolic helix in repeat II (designated the CH2II helix) is deleted. This mutant, named ΔCH2, still reserves a down VSDII, but PIP2 is invisible and the juxtamembrane region on the cytosolic side is barely discernible. Our structural and electrophysiological characterizations of the wild type and ΔCH2 Cav2.3 show that the CH2II helix stabilizes the inactivated conformation of the channel by tightening the cytosolic juxtamembrane segments, while CH2II helix is not necessary for locking the down state of VSDII.
Similar content being viewed by others
Introduction
Voltage-gated calcium (Cav) channels permeate Ca2+ influx in response to the membrane depolarization. They regulate a broad range of physiological processes, such as muscle contraction, neurotransmitter release, hormone secretion, and cell death1,2,3,4. The 10 primary subtypes of mammalian Cav channels are divided into three subfamilies, Cav1, Cav2, and Cav3, based on the phylogeny of the core α1 subunits. Cav1 and Cav3 channels are also known as the L-type and T-type channels, respectively, for their distinct electrophysiological properties. The three Cav2 members, known as the P/Q-type for Cav2.1, the N-type for Cav2.2, and the R-type for Cav2.3, play an important role in signal transduction in the central and peripheral nervous systems1,2,3,4,5.
A systematic structural analysis of Cav channels will not only reveal the molecular basis for their working principles, but also facilitate drug discovery targeting various Cav channelopathies4,6,7. Benefiting from the resolution revolution of single-particle cryo-electron microscopy (cryo-EM), we have solved the high-resolution structures of representative members from each Cav subfamily, Cav1.1, Cav1.3, Cav2.2, and Cav3.1 over the years since 20158,9,10,11,12. These structures reveal the common architecture of Cav channels. The transmembrane regions of the α1 subunits all share the canonical voltage-gated ion channel fold, wherein four homologous repeats (designated I-IV), each containing six transmembrane helices (S1-S6), are interwoven in a domain-swapped manner13,14. The four sets of S5 and S6 segments enclose the central pore domain (PD), which is responsible for the selective Ca2+ permeation. The S1-S4 segments in each repeat constitute the voltage sensing domains (VSD) that undergo conformational shifts in response to membrane potential changes, transmitting the electric signals to pore gating15,16,17,18.
More rewarding are structural discoveries of the subtype-specific features. Of particular note, an endogenous phosphatidylinositol 4,5-bisphosphate (PIP2) molecule is seen to attach to the interface of VSDII and the PD on the inner membrane leaflet of human Cav2.211,19. In addition, the exceptionally long S6II segment is followed by two consecutive helices, designated CH1II and CH2II, which fold back toward the membrane. A Trp residue that marks the amino terminus of CH2II appears to play a critical role to secure the closed state of the intracellular gate11,19. To investigate whether these structural features are unique to Cav2.2 or conserved in the Cav2 subfamily, we sought to solve the structures of human Cav2.1 and Cav2.3. As of now, we have not been able to produce suitable samples of Cav2.1 for cryo-EM analysis. Our present study focuses on Cav2.3.
Cav2.3 channels, a ternary complex comprising the extracellular α2δ and the cytosolic β subunits in addition to the α1 subunit, are strongly expressed in the cortex, hippocampus, striatum, amygdala, and interpeduncular nucleus20. Six different splice variants of the α1 subunits, Cav2.3a-f, have been identified in various mammalian species21. Dysfunction of Cav2.3 is the major cause of developmental and epileptic encephalopathy 69 (DEE69), a severe encephalopathic disorder characterized by refractory seizures and neurodevelopmental impairment22. Cav2.3-deficient mice display reduced seizure activity, altered pain response, and protection from Parkinson’s disease-related neurodegeneration23,24,25. Therefore, Cav2.3 channels represent potential drug targets for managing epileptic seizures and neurological disorders26.
Unlike the P/Q-type and the N-type Cav channels, the Cav2.3 channels display low sensitivities to ω-conotoxins, a group of neurotoxic peptides that have been used as tools to explore the physiological functions of Cav2 channels27,28. Cav2.3 channels are subject to selective inhibition by SNX-482, a 41-aa peptide toxin from the tarantula Hysterocrates gigas, which exhibits an antinociceptive activity26. Furthermore, the Cav2.3 channels show a more rapid inactivation and slower recovery from inactivation, which contributes to their specific functional roles in neurons29.
Here, we report the structural analysis of the human Cav2.3 complex. The high-resolution structure provides an accurate template for drug discovery. To investigate the role of the CH2II helix, we determined the cryo-EM structures of a CH2II-truncated mutant (named as ΔCH2) in addition to the full-length channel. Our studies show that CH2II stabilizes the inactivated conformation of the channel by tightening the juxtamembrane region on the cytosolic side.
Results
Nearly identical structure of full-length Cav2.3 with Cav2.2
The full-length human Cav2.3 ternary complex consisting of α1, α2δ−1, and β3 was recombinantly co-expressed, purified, and analyzed with cryo-EM using a protocol nearly identical to that for Cav2.211. The only modification is the addition of 220 μM SNX-482, that was reported to specifically inhibit Cav2.3 by binding to VSDs III and IV26,30. However, there was no discernible density for the peptide toxin throughout data processing. It is possible that the purification condition, which disrupts the membrane potential, is incompatible with SNX-482 binding, as depolarization was reported to reverse toxin association30.
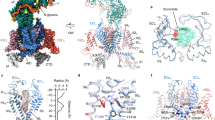
A 3D EM reconstruction for the Cav2.3 ternary complex was determined at an average resolution of 3.1 Å (Fig. 1a, b, Supplementary Figs. 1, 2 and Supplementary Table 1). As seen in other Cav channel complexes9,11, α1 and α2δ−1 subunits are well resolved, while β3 is of lower resolution. The structure of β3 from the Cav2.2 complex (PDB: 7MIY) was docked as a rigid body.
a Heat map for the local resolutions of the Cav2.3 EM reconstruction. The map, calculated in RELION, was generated in ChimeraX. b 3D EM map of human Cav2.3 complex comprising the extracellular α2δ−1 subunit (light pink for α2, and green for δ), the transmembrane α1 subunit (domain-colored), and the cytosolic β3 subunit (salmon). The cytosolic helix of repeat II, designated as CH2II, is colored chocolate. Sugars and lipids are colored wheat and orange, respectively. Also labeled are the α1-interacting domain (AID) and the carboxy-terminal domain (CTD). The same color scheme is applied to the structure of full-length Cav2.3 in the manuscript unless otherwise indicated. c Nearly identical structures of Cav2.2 and Cav2.3. The structures of Cav2.2 (purple, PDB code 7MIY) and Cav2.3 (domain-colored) can be superimposed with the root-mean-square deviation (RMSD) of 0.81 Å for 1905 Cα atoms in the α1 and α2δ−1 subunits. Due to the low resolution, the β3 structure from the Cav2.2 complex was docked as a rigid body in the map of Cav2.3. PIP2 (black sticks) binds at the interface of VSDII and the PD of human Cav2.3 in the inner membrane leaflet.
The overall structure of Cav2.3 is nearly identical to that of apo-Cav2.2, with the root-mean-square deviation (RMSD) of 0.81 Å for 1905 Cα atoms in the α1 and α2δ−1 subunits (Fig. 1c). Briefly, the α1 subunit is also featured with a down VSDII, while the other three VSDs are in the typical depolarized or up conformations (Supplementary Fig. 3). The closed intracellular gate of PD is secured by CH2II. And a PIP2 molecule bound to the same position is also clearly resolved (Fig. 1c and Supplementary Fig. 2b). Five PIP2 species, 34:1, 36:2, 36:1, 38:3, and 38:2, were detected in the lipidomic analysis of purified channel complex (Supplementary Fig. 2c). These observations suggest that the structural and functional roles of CH2II and PIP2 may be conserved in the N- and R-type Cav channels. The shared structural features of Cav2.2 have been thoroughly depicted in our previous report11. Here we will avoid redundant structural illustrations, but focus on CH2II for detailed analysis.
CH2II stabilizes an inactivated state of the channel
Cav2.3 has a similar voltage dependence for activation and steady-state inactivation to Cav2.2, with V1/2 measured at −14.7 ± 0.3 mV and −70.9 ± 0.1 mV, respectively (Fig. 2a and Supplementary Figs. 4a–c). When the holding potential was set at −100 mV, more than 90% of the Cav2.3 channels activated at 0 mV (Fig. 2a). But when the holding potential was at −40 mV or more depolarized, most channels were trapped in the inactivated state (Fig. 2a). As the recombinant channels were expressed in HEK293F cells and purified at 0 mV, the purified channels are likely trapped in an inactivated state. As briefly mentioned in the introduction, a Trp residue at the beginning of CH2II serves as an organizing center for intracellular gating residues. In analogy, it is like a bolt that tightens the S6 tetrahelical bundle at the intracellular gate (Fig. 2b).
a Deletion of the entire CH2II segment (mutant named as ΔCH2) did not affect channel activation, but led to a pronounced right shift in the steady-state inactivation. Half-maximal activation: V1/2 (WT) = −14.7 ± 0.3 mV, V1/2 (ΔCH2) = −12.3 ± 0.2 mV; Half-maximal inactivation: V1/2 (WT) = −70.9 ± 0.1 mV, V1/2 (ΔCH2) = −57.3 ± 0.1 mV. All data are presented as mean ± SEM. The number of tested cells is labeled in parentheses. Source data are provided as a Source Data file. b CH2II (colored chocolate) is positioned right beneath the intracellular gate and surrounded by multiple intracellular segments. Inset: CH2II tightens the closed intracellular gate. Trp778 (chocolate sphere) at the beginning of CH2II helix interacts with gating residues (domain-colored sticks) on the S6 tetrahelical bundle. c ΔCH2 mutant recovers from inactivation much faster than WT channel. The electrophysiological protocols are shown above the curves. After treating with pre-pulses at −40 mV (left) or +10 mV (right) for 300 ms to induce inactivation, −100 mV recovery voltage was applied for variable times (0-1000 ms), which is then followed by a 30-ms test pulse at +10 mV. For pre-pulse at −40 mV, the fitted time constants (τ) are 90.3 ± 5.4 ms and 33.9 ± 1.3 ms for WT and ΔCH2, respectively. For pre-pulse at +10 mV, τ = 137.4 ± 4.0 ms for WT and 40.0 ± 2.0 ms for ΔCH2. Source data are provided as a Source Data file.
Supporting the structural analysis, deletion of the entire CH2II helix (residues 773-791, the resulting mutant named ΔCH2) had no impact on channel activation, but shifted the V1/2 for the steady-state inactivation from −70.9 ± 0.1 mV to −57.3 ± 0.1 mV (Fig. 2a and Supplementary Fig. 4c). We then measured the rates of recovery for WT and ΔCH2 treated at different pre-pulses. For these experiments, pre-pulses of −40 mV or 10 mV were respectively applied for 300 ms to induce inactivation. After holding the cells at the hyperpolarization of −100 mV for variable durations, the channel opening at 10 mV was recorded (Fig. 2c and Supplementary Fig. 4d). Supporting the role of CH2II to secure the inactivated state, ΔCH2 mutant recovered faster than WT channel under both settings (Fig. 2c). For recovery from the pre-holding at −40 mV, the fitted time constants (τ) of WT and ΔCH2 channels are 90.3 ± 5.4 ms and 33.9 ± 1.3 ms, respectively (Fig. 2c, left). When pre-holding was set at 10 mV, ΔCH2 also accelerated the recovery with τ shortened from 137.4 ± 4.0 ms to 40.0 ± 2.0 ms (Fig. 2c, right). The electrophysiological characterizations consolidate the structural analysis that the CH2II helix stabilizes the inactivated conformation of Cav2.3.
As CH2II is unique to Cav2 channels and only Cav2.2 and Cav2.3 show a down VSD among all resolved Cav structures, we next examined if ΔCH2 would display a similar conformation to the structures of Cav1.1 and Cav1.3, in which all four VSDs are up9,12.
The juxtamembrane domains are less ordered in ΔCH2
The protein expression level and solution behavior of ΔCH2 are similar to that of the WT channel complex. Following our standard protocol, the structure of ΔCH2 was determined at an overall resolution of 3.1 Å out of 68,109 particles (Supplementary Fig. 5). Despite a similar nominal resolution for the overall structure to that of the WT channel, several functional units were only poorly resolved in ΔCH2.
In the absence of CH2II, the segments on the intracellular border of the membrane and in the cytosol, including the α1-interacting domain (AID), which is the elongated helix succeeding S6I, the cytosolic fragment of S6II, the III-IV linker and the carboxy-terminal domain (CTD), are barely discernible in the final map (Fig. 3a). The entire β3 subunit is completely invisible. Within the transmembrane region, VSDII and VSDIV display lower local resolutions, especially for their S2 and S3 segments (Fig. 3b). These observations support that CH2II stabilizes the intracellular side of the juxtamembrane region of the channel.
a Almost all the intracellular segments, including the β3 subunit, the AID, the cytosolic fragment of S6II, and the CTD are barely discernible in the map of ΔCH2. The unresolved segments are highlighted in the map of full-length channel (WT) with the same color scheme as in Fig. 1b. For visual clarity, both maps are low pass-filtered to 6 Å. b VSDII (top) and VSDIV (bottom) display discontinuous densities for the S2 and S3 segments in the absence of the CH2II helix. c PIP2 is not resolved in ΔCH2 map. The density for PIP2, colored pink in the WT map, is missing in the map of ΔCH2.
Of particular note, there is no density for PIP2 in the 3D reconstruction of ΔCH2 (Fig. 3c). To distinguish if the invisibility of PIP2 is due to loss of binding or poor resolution associated with local structural flexibility, we characterized the presence of PIP2 in WT and ΔCH2 using lipidomic analysis. The abundance of PIP2 indeed dropped in ΔCH2 by about a half (Supplementary Fig. 6).
CH2II couples PD gating
Next, we compared the structures of ΔCH2 and WT Cav2.3 for detailed analysis. A total of 1939 out of 3416 side chains were assigned for α2δ−1 and the extracellular and transmembrane region of α1 in ΔCH2 (Supplementary Fig. 7 and Supplementary Table 1). For the α1 segments with lower resolutions, such as the S3 segments in VSDII and VSDIV, poly-Ala were assigned. The overall structures of the α1 subunit of ΔCH2 and WT Cav2.3 can be superimposed with a RMSD of 0.57 Å over 775 Cα atoms (Fig. 4a). We had expected that VSDII would exhibit an “up” conformation; however, it is clear that VSDII in ΔCH2 remains down, identical to that in the WT channel (Supplementary Fig. 8a). In contrast, the ensuing S4-5II and the PD exhibit evident structural deviations (Fig. 4a and Supplementary Fig. 8b).
a In the superimposed α1 structures of WT and ΔCH2 Cav2.3, S6I, S6II and S4-5II segments exhibit evident conformational changes. Blue arrows indicate the structural shifts from WT to ΔCH2. b The gating residues on S6I displace outwards (left), while those on S6II undergo an axial rotation (middle). The structural shifts of gating residues from WT to ΔCH2 are indicated by red arrows. A secondary structure transition from π → α occurs in S6II. Right: In the absence of CH2II, the intracellular gate becomes loosely closed. The pore radii are calculated by HOLE53. Source data are provided as a Source Data file. c Opening of the I-II (left) and II-III (right) fenestration sites upon the axial rotation of S6II that turns Tyr692 and Phe698 (labeled red) away in ΔCH2. d Rotations of Asp704 and Asn708 on S6II induce the upward motion of S4-5II and its joint with S5II. Purple dashes indicate the H-bonds between Ser588 and Ser589 on S4-5II with Asp704 and Asn708 on S6II. Structural shifts of the interacting residues are indicated by black arrows. Right: The hinge between S4-5 and S5II moves toward the membrane by a displacement of ~ 6 Å. e The local shift of S4-5II segment disfavors PIP2 binding. The upward movement would lead to potential clash with PIP2 binding.
Both S6I and S6II slightly move outwardly, resulting in a loosened but still closed intracellular gate (Fig. 4b). Accompanying the rotation of S6I and S6II, the previously sealed I-II and II-III side walls of the PD now both have fenestrations in ΔCH2 (Fig. 4c). Therefore, the PD in the WT Cav2.3 is in a tightly closed state, whereas that in ΔCH2 is loosely closed, as seen in the structures of ligand-free Cav1.1, Cav1.3,and Cav3.19,10,12. Consistent with previous structural observations in Cav and Nav channels31, S6II has one π-helical turn in the tightly closed WT channel, which transforms to the α conformer in the ΔCH2 structure (Fig. 4b, c).
Rotation of S6II places two polar residues, Asp704 and Asn708, to face S5II and S4-5II. In the structure of ΔCH2, the S4-5II segment and its hinge with S5II undergo the most pronounced structural shift; the hinge rises towards the membrane by approximately 6 Å. In this conformation, Asp704 on S6II can form hydrogen bonds (H-bonds) with Ser598 and Ser599, and Asn708 is also H-bonded to Ser598 (Fig. 4d). A lifted hinge between S4-5II and S5II together with an unchanged VSDII result in a sharpened angle between S4II and S4-5II, which is no longer compatible with PIP2 binding. This conformational change thus explains the lack of PIP2 density in the map of ΔCH2 (Fig. 4d, e and Supplementary Fig. 8b).
Distinct conformations of AID in ΔCH2
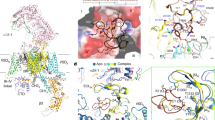
Although the intracellular region is invisible in the final high-resolution map, there are still densities in the low pass-filtered map. We attempted to probe the structural heterogeneity of ΔCH2 using 3D variability analysis (3DVA) in cryoSPARC32. Potential motion trajectories were analyzed in multi-dimensional conformational space, in which one mode shows a significant dynamic motion of the AID, CTD, and VSDIV (Fig. 5, Supplementary Fig. 5d and Supplementary Movie 1).
a Two extreme trajectories from 3DVA reveal a “transverse” AID (1st frame, designated as ΔCH2-AIDt) versus a “straight” AID (10th frame, designated as ΔCH2-AIDs). These two reconstructions are low pass-filtered to 7 Å. b The α1 subunit in ΔCH2-AIDt (wheat) and WT Cav2.3 (gray) can be well aligned. c Accompanying the straightening of AID, CTD and VSDIV also undergo a concerted swing. Red arrows indicate the structural shifts from ΔCH2-AIDt (wheat) to ΔCH2-AIDs (light blue). Also referred to Supplementary Movie 1 for the conformational shifts.
Two extreme trajectories, the 1st (designated as ΔCH2-AIDt) and the 10th (ΔCH2-AIDs) frames of the reconstructed volume series, together with the full transition along the volume series reveal a marked conformational shift of the AID from a WT-like “transverse” conformation to a “straight” one (Fig. 5a, b and Supplementary Movie 1). Meanwhile, the CTD undergoes a concerted swing accompanying the straightening of the AID (Fig. 5c and Supplementary Movie 1). VSDIV, particularly its S1IV segment that is directly connected to the III-IV linker that binds to the CTD, moves along the same direction but to a smaller degree than the CTD (Supplementary Movie 1).
Discussion
Our present study set out to address the following questions. 1) Is the Cav2-unique CH2II helix the determinant for the down state of VSDII in the cryo-EM structure? 2) What are the functional and structural roles of the CH2II helix? Our structural and electrophysiological characterizations of human Cav2.3 confirm the role of the CH2II helix in stabilizing the inactivated conformation of the channel. In the structure of CH2II-deleted Cav2.3, the PD is loosely closed, but VSDII remains down. The overall conformation is no longer compatible with PIP2 binding. Therefore, the structure of ΔCH2 demonstrates that the CH2II helix is not necessary for the down state of VSDII. Sequence analysis cannot offer an immediate clue to the determinant for the down state of VSDII. Future characterizations, such as engineering of Cav1 and Cav2 chimera followed by the systematic mutagenesis, may unveil the determinants for the conformational switch of VSDII.
An unexpected discovery is the conformational flexibility of the AID. In all the reported structures of Cav1.1, Cav1.3, and Cav2.28,9,11,12,19,33,34, AID is a transverse helix lying on the intracellular surface of the membrane, connected to S6I through a short turn. 3DVA of the low pass-filtered map of ΔCH2 reveals a potentially straightened conformation of the AID as a natural extension of S6I into the cytosol. When the maps were further low pass-filtered to ~15 Å, additional densities on the cytosolic side were shown (Supplementary Fig. 9). In reference to the low pass-filtered map for WT Cav2.3, this density likely belongs to the β3 subunit, which is invisible in the high-resolution map (Fig. 3a and Supplementary Fig. 9). The position and contour of the density in ΔCH2-AIDt are similar to that in the WT map (Supplementary Fig. 9, upper row). In the map of ΔCH2-AIDs, however, the density appears to undergo a large degree of rotation (Supplementary Fig. 9, lower row). If this density indeed corresponds to β3 that binds to AID in both bent and straightened conformation, the shift of the density is consistent with the shift of AID. But as the density can only be seen in the low pass-filtered maps analyzed with 3DVA, we cannot rule out the possibility that this density may not belong to β3.
In light of the potential conformational diversity of AID and β3 in ΔCH2, we re-examined the maps of WT Cav1.1, Cav2.2, and Cav2.3 (EMDB codes: EMD-22426, EMD-23868, and EMD-28529, respectively). In the presence of CH2II, the β3 subunit is sandwiched between the transverse AID and the elongated S6II in Cav2.2 and Cav2.3. There is no additional conformation for the AID. In Cav1.1, which does not possess a CH2II helix, AID and the bound β1 do show certain conformational heterogeneities, but AID remains to be bent9. Of note, there is one major difference between WT Cav1.1 and Cav2.3-ΔCH2. VSDII, which interacts with the bent AID, is up in Cav1.1 and down in Cav2.3-ΔCH2. Considering that the β1 subunit is critical for the coupling between Cav1.1 and the downstream RyR1 in skeletal muscle35,36,37, the dramatic conformational shift of AID indicated by the 3DVA analysis immediately raised the question of whether AID is straightened in a resting state Cav1.1 channel, wherein VSDII is down, a speculation to be tested in the future.
Cav2.3 channels represent potential targets for the treatment of various neurological diseases22,38. However, there have lacked effective and selective Cav2.3 modulators as of today. Our structures provide an accurate molecular template for mapping disease mutations (Supplementary Fig. 10) and may facilitate the structure-based drug design22. Our structure-based discoveries serve as a framework to probe the electromechanical coupling of Cav channels and shed light on the mechanistic investigation of the excitation-contraction coupling of skeletal muscles that involve Cav1.1 and β1.
Methods
Transient expression of human Cav2.3 in HEK293F cells
Codon-optimized cDNAs of CACNA1E for full-length Cav2.3 α1 (2,313 residues, Uniprot Q15878-1), CACNA2D1 for α2δ−1 (1,103 residues, Uniprot P54289-1) and CACNB3 for β3 (484 residues, Uniprot P54284-1) were synthesized (BGI Geneland Scientific). For CH2-deleted Cav2.3 (ΔCH2), residues 773-791 of the α1 subunit were deleted with standard two-step PCR. All the subunits were cloned into the pCAG vector, with an amino-terminal Flag-tag and a carboxy-terminal His10-tag at the α1 and β3 subunits. For western blotting to examine the stoichiometry of the α1 and β3 subunits, β3 subunit was cloned into the pCAG vector with amino-terminal twin strep-tag. HEK293F suspension cells (Thermo Fisher Scientific, R79007) were cultured in Freestyle 293 medium (Thermo Fisher Scientific) at 37 °C, supplied with 5% CO2 under 60% humidity. When cell density reached 1.5-2.0 × 106 cells per mL, a mixture of expression plasmids including 0.75 mg α1, 0.6 mg α2δ−1 and 0.5 mg β3, and 3 mg polyethylenimine (Polysciences) were added into the cell culture for transient expression of human Cav2.3 complex.
Protein purification of human Cav2.3
For one batch of protein purification, 8 L of HEK293F cells were collected approximately 72 h after transfection by centrifugation at 3600 g for 10 min and resuspended in the lysis buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM CaCl2 and the protease inhibitor cocktail containing 2.6 μg mL−1 aprotinin (VWR Life Science) and 1.4 μg mL−1 pepstatin (VWR Life Science). The suspension was supplemented with glycol-diosgenin (GDN, Anatrace) to a final concentration of 1% (w/v), n-dodecyl-β-D-maltopyranoside (DDM, Anatrace) to 0.2% (w/v), and cholesteryl hemisuccinate Tris salt (CHS, Anatrace) to 0.04% (w/v). After incubation at 4 °C overnight, the mixture was centrifuged at 35,000 g for 30 min, and the supernatant was applied to anti-Flag M2 affinity resin (Sigma) for affinity purification. The resin was rinsed with wash buffer (buffer W) containing 25 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM CaCl2, and 0.01% GDN and eluted with buffer W supplemented with 0.2 mg mL−1 Flag peptide (synthesized by GenScript). The eluent was then concentrated using a 100-kDa molecular weight cut-off Amicon filter unit (Millipore) and further purified through size-exclusion chromatography (Superose 6 10/300 GL, GE Healthcare) that was pre-equilibrated in buffer W. The peak fractions were pooled and concentrated to a final concentration of about 20 mg mL−1 with α2δ−1 in excess.
For structural determination of Cav2.3 in the complex with peptide toxin, purchased SNX-482 (Alomone labs) was only added to the concentrated wild-type protein solution at a final concentration of 220 μM. The mixture was incubated at 4 °C for 30 min before making cryo-grids.
For western blotting, 1.5 L of HEK293F cells expressing the wild-type or ΔCH2 mutant Cav2.3 proteins were purified following the same procedure. The eluents after anti-Flag resin were separated by SDS-PAGE and transfected onto PVDF membrane (Millipore). The membranes were blocked by 5% (w/v) nonfat milk (Bio-Rad) in TBST buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, and 0.05% (w/v) Tween-20. The membranes were incubated with primary monoclonal anti-Flag® M2 antibody (1:3000 dilution, Sigma-Aldrich) against the α1 subunit or Strep Tag II monoclonal antibody (1:2500 dilution, Invitrogen, clone 1810CT579.47.56.10) against the β3 subunit, and IRDye® 800CW goat anti-mouse IgG secondary antibody (1:4000 dilution, Li-COR). The membranes were exposed by Odyssey® CLx imaging system (LI-COR 9140).
Cryo-EM sample preparation and data collection
Aliquots of 3.5 μl concentrated WT or ΔCH2 Cav2.3 proteins were loaded onto glow-discharged holey carbon grids (Quantifoil Cu/Au R1.2/1.3, 300 mesh), which were blotted for 6 s and plunge-frozen in liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV (Thermo Fisher) at 8 °C with 100% humidity. Grids were transferred to a Titan Krios electron microscope (Thermo Fisher) operating at 300 kV and equipped with a Gatan Gif Quantum energy filter (slit width 20 eV) and spherical aberration (Cs) image corrector. Micrographs were recorded using a K2 Summit counting camera (Gatan) in super-resolution mode with a nominal magnification of 105,000×, resulting in a super-resolution pixel size of 0.557 Å. Each stack of 32 frames was exposed for 5.6 s, with an exposure time of 0.175 s per frame. The total dose for each stack was about 50 e− per Å2. The dose rate is 10.6 e-/ pixel/s. SerialEM was used for fully automated data collection39. All 32 frames in each stack were aligned, summed, and dose-weighted using MotionCorr240 and twofold-binned to a pixel size of 1.114 Å per pixel. The defocus values were set from −1.9 to −2.1 μm and estimated by Gctf41.
Image processing
For WT Cav2.3, the images were collected in three batches. Although the last two batches of samples were added SNX-482, no density for the drug was observed. A total of 7,160 (1st: 2,568; 2nd: 2,245; 3rd: 2,347) cryo-EM micrographs were collected and 1,664,705/1,962,632/2,117,770 particles were auto-picked by RELION-3.042. Particle picking was performed using 2D classes of human Cav2.2 (EMD-23868) in the side and tilted views as reference. All subsequent 2D and 3D classification and refinement were performed with RELION-3.0. One round of reference-free 2D classification was performed to remove ice spots, contaminants, and aggregates, yielding 1,415,239/1,692,632/2,072,104 particles. The particles were then processed with the global search 3D multi-reference classification with K = 4 using bin2 particles. The EM map of human Cav2.2 (EMD-23868)11, low pass-filtered to 20 Å, was used as an initial good reference. The output of 1st and 2nd datasets were applied to the local angular search 3D classification with four classes. The 3rd dataset was performed skip align 3D classification. A total of 221,390/345,032/50,314 particles were selected by combining the good classes. The particles were then re-extracted using a box size of 280 pixels and pixel size of 1.114 Å. These particles yielded reconstructions at 3.7/4.5/3.7 Å after 3D auto-refinement with an adapted mask. Skip align 3D classification for 1st and 2nd datasets using bin1 particles and Bayesian polishing for all datasets resulted in reconstructions at 3.2/3.6/3.5 Å from 47,971/64,770/50,314 particles. Skip align 3D classification for the merged particles from three datasets afforded the final reconstruction at 3.1 Å out of 118,244 particles.
For ΔCH2, A total of 6,160 (1st: 1,930; 2nd: 2,563; 3rd: 1,667) cryo-EM micrographs were collected and 1,530,824/1,696,892/1,236,150 particles were auto-picked. One round of reference-free 2D classification yielded 1,500,417/1,535,373/1,123,979 particles. The particles were then processed with a global search 3D multi-reference classification with K = 4 using bin2 particles. The particles of 1st dataset were then applied to skip align 3D classification, and the output of 2nd and 3rd datasets were then processed with local angular search 3D classification. A total of 55,655/329,382/184,237 particles were selected by combining the good classes. Bin1 particles yielded reconstructions at 4.4/4.2/4.3 Å after 3D auto-refinement with an adapted mask. Skip align 3D classification for 2nd dataset only and Bayesian polishing for all datasets resulted in reconstructions at 4.2/3.2/3.6 Å from 55,655/38,415/30,126 particles. Skip align 3D classification for the merged particles yielded the reconstruction at 3.1 Å out of 68,109 particles. For WT and ΔCH2, > 97% of junk particles and low-resolution particles were thrown out during data processing.
All 2D classification, 3D classification, and 3D auto-refinement were performed with RELION 3.0. Resolutions were estimated using the gold-standard Fourier shell correlation 0.143 criterion with high-resolution noise substitution43,44.
Model building and refinement
Model building for human Cav2.3 used Cav2.2 complex comprising α1, α2δ−1, and β3 subunit (PDB code 7MIY) as the starting model. The structure of Cav2.2 was docked to the Cav2.3 map using UCSF Chimera45 and then manually adjusted in COOT46. The lipids were manually built to fit into the corresponding densities in the map. The model was refined against the corresponding map by the phenix.real_space_refine program in PHENIX47 with secondary structure and geometry restraints. For ΔCH2 structure, the model was manually adjusted and refined in COOT based on the WT structure. Statistics of 3D reconstruction and model refinement can be found in Supplementary Table 1. All structure figures were prepared in UCSF Chimera45, ChimeraX48 and PyMol49.
3D variability analysis of CH2II-deleted structure (ΔCH2)
The 3D variability analysis was performed in cryoSPARC32,50. ΔCH2 particles after Bayesian polish (in total 124,196 particles) were applied for one round 2D classification. 8,161 junk particles were used to generate bad references in 3D ab-initio reconstruction program. 116,035 particles were selected and applied to heterogeneous refinement using 1 good and 2 bad references. The selected 90,380 particles afforded a 3.1 Å reconstruction after non-uniform refinement. The output from non-uniform refinement was applied to 3D variability analysis with three components. Initial results were processed as a simple mode for preview. A set of different subset numbers for intermediate reconstruction were tested and 10 was selected and applied for the final presentation. The first and last frames together with the high-resolution structure of ΔCH2 were used for heterogeneous refinement of the 90,380 particles, resulting in a similar reconstruction as shown in intermediate mode with the portions of particle for bend state (13%) and straight state (14%), respectively.
Whole-cell voltage-clamp recordings
HEK293T cells (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% (v/v) fetal bovine serum (PAN-Biotech) at 37 °C with 5% CO2. For 35 mm culture dish, 0.5 μg cDNA of each subunit, 0.5 μg pIRES2-EGFP (Clontech) plasmid, and 6 μg polyethylenimine (PEI) (Polysciences) were used for transfection. Whole-cell voltage-clamp electrophysiology was performed 36 to 60 hours after transfection at room temperature. Isolated, GFP-positive cells were then selected for whole-cell recordings.
The extracellular solution contained 160 mM TEA-Cl, 1 mM BaCl2, 1 mM CaCl2, and 10 mM HEPES (pH 7.3). The intracellular solution contained 140 mM CsCl, 10 mM HEPES, and 10 mM EGTA (pH 7.4). Glass pipette electrodes with a resistance of 2-5 MΩ were used. Whole-cell currents were acquired using a MultiClamp 700B Amplifier and a Digidata 1550B digitizer with pCLAMP 10 software (Molecular Devices). Data were collected at a 20 kHz sample rate and filtered at 5 kHz with a low-pass filter. The series resistance was ~5-10 MΩ and was compensated ~80–90%.
To obtain the activation curves of WT and ΔCH2 Cav2.3 channels, cells were held at −100 mV followed by a series of 100-ms voltage steps from −60 mV to +50 mV in 5 mV increments. Data analyses were performed using Clampfit (Molecular Devices) and GraphPad Prism 7 (GraphPad Software). The currents of Cav2.3 were converted into conductance using the following equation:
where g represents conductance, I for Cav2.3 current, V for tested membrane potential, and Vrev for reversal potential. The conductance data were then fitted with the Boltzmann equation below to generate the steady-state activation curve:
Here g is the conductance at a test voltage, gmax is the maximal conductance of Cav2.3 across tested voltages, V is the tested voltage, Vmid is the half-maximal activation voltage, and k is the slope factor.
To generate the steady-state inactivation curve, cells were held at −100 mV followed by a series of 1 s voltage steps from −100 mV to +20 mV in 5 mV increments (pre-pulse), followed immediately by a 50 ms test pulse to +10 mV.
The steady-state inactivation curves were then fitted with the Boltzmann equation below:
Here I is the current during the test pulse after preconditioning at a certain voltage, Imax is the maximal current of Cav2.3, V is the tested pre-pulse voltage, Vmid is the half-maximal inactivation voltage, and k is the slope factor.
To test the time-dependent recovery from inactivation, cells were initially held at −100 mV and depolarized to either +10 mV or −40 mV for 300 ms to inactivate channels. A recovery hyperpolarization step to −100 mV was then applied for a variable period (0, 5, 10, 25, 50, 75, 100, 125, 150, 200, 300, 500, 800, and 1000 ms), followed by a 30 ms test pulse to +10 mV. Currents at the test pulse were collected and plotted over the recovery time. The curves showing recovery from inactivation were fitted using the single exponential equation below:
Here I is the current at the test pulse after a certain recovery time, Imax is the current at 1000 ms recovery, t is the recovery time, and τ is the time constant of recovery from the inactivated state.
Lipidomic analysis of PIP2
Protein samples were buffer-exchanged to 1 M ammonium acetate, pH 7.0 with 0.0042% GDN, and digested with trypsin overnight at 37 °C. The peptide/lipid mixture was dried using a SpeedVac vacuum concentrator (Thermo Fisher Scientific) and dissolved in 70% mobile phase A (acetonitrile/H2O: 60/40, 10 mM ammonium formate and 0.1% formic acid) and 30% mobile phase B (isopropanol/acetonitrile: 90/10, 10 mM ammonium formate and 0.1% formic acid). The lipids were loaded onto a C8 column (Acclaim PepMap 100, C8, 75 µm × 15 cm, Thermo Scientific) by a Dionex UltiMate 3000 RSLC Nano system coupled to an Eclipse Tribrid mass spectrometer (Thermo Scientific). The lipids were separated with a gradient from 20% to 70% mobile phase B. Typical MS settings were spray voltage of 2.2 kV and heated capillary temperature of 320 °C. For data-dependent acquisition, full MS scans were acquired in the Orbitrap (m/z 300-2000) with a resolution of 120000 in negative ion mode. Fragment spectra were acquired in the Orbitrap with a resolution of 15000 using higher-energy collisional dissociation (HCD) with stepped collision energies (25% and 30%).
The raw LC-MS/MS data were converted to mgf format and processed with LipiDex (v1.1) for phospholipid identification using LipiDex_HCD_Formic and a manually curated glycolipid library51. The MS and MS/MS search tolerances were set to 0.01 Th. Phospholipid quantification was also performed using LipiDex. The peak features from raw LC-MS/MS data were firstly extracted using MZmine (v.2.53)51 with noise level of 5 × 104. The chromatograms were integrated using local minimum search algorithm with minimum absolute height of 5 × 105 and peak duration range of 0.05-1.50 min. The isotopic peaks were grouped and aligned with m/z tolerance of 0.005 Th/10.0 ppm and retention time tolerance of 0.5 min. The quantified peak features were matched to the identified lipid species with minimum MS2 search dot product of 500 and MS2 search reverse dot product of 700. The quantified lipids were manually examined and normalized for relative quantification.
For phosphoinositides analysis, the LC-MS method was adapted from Ogsio et al. and tailored for identifying co-purified lipids with membrane proteins52. Briefly, the protein samples were buffer-exchanged to 1 M ammonium acetate, pH 7.0 with 0.0042% GDN, and digested with trypsin overnight at 37 °C. The peptide/lipid mixture was dried and dissolved in 100% methanol. No phosphoinositide enrichment was performed before LC-MS analysis. The samples were diluted to 50% methanol with water, just before LC-MS analysis. The phosphoinositides were analyzed on a C8 column (PepMap 100, C8, 75 µm × 15 cm, Thermo Fisher Scientific) held at 35 °C using a Dionex UltiMate 3000 RSLC Nano system. A binary buffer system was applied to separate phosphoinositides. Mobile phase A was methanol/water/ethylamine (50/50/0.1) and mobile phase B was isopropanol/ethylamine (100/0.1). The lipids were separated with a liner gradient from 5 % to 90% mobile phase B at a flow rate of 300 nl/min. The LC system was coupled to an Eclipse Tribrid mass spectrometry in negative ion mode (Thermo Scientific). For data-dependent acquisition, full MS scans were acquired in the Orbitrap (m/z 300-1800) with a resolution of 120000. Fragment spectra were acquired in the Orbitrap with a resolution of 15000 using HCD with stepped collision energy (25% and 30%). The raw data was processed manually for phosphoinositide identification and quantification.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon reasonable request. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under the accession codes EMD-28529 (wild-type Cav2.3) and EMD-28530 (ΔCH2 mutant Cav2.3). The coordinates have been deposited in the RCSB Protein Data Bank (PDB) under the accession codes 8EPL (wild-type Cav2.3) and 8EPM (ΔCH2 mutant Cav2.3). The proteins for structural comparison in this study can be found in PDB under the accession code 7MIY (human Cav2.2), and in EMDB under the accession codes EMD-22426 (rabbit Cav1.1) and EMD-23868 (human Cav2.2). The lipidomic raw data have been deposited on Figshare (https://doi.org/10.6084/m9.figshare.21502188). The source data underlying Figs. 2a, c, 4b and Supplementary Figs. 1b, 4a, b, 5a, 6a, c are provided as a Source Data file. Source data are provided with this paper.
References
Catterall, W. A. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev. Cell Dev. Biol. 16, 521–555 (2000).
Nanou, E. & Catterall, W. A. Calcium Channels, Synaptic Plasticity, and Neuropsychiatric Disease. Neuron 98, 466–481 (2018).
Zamponi, G. W. Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat. Rev. Drug Disco. 15, 19–34 (2016).
Zamponi, G. W., Striessnig, J., Koschak, A. & Dolphin, A. C. The Physiology, Pathology, and Pharmacology of Voltage-Gated Calcium Channels and Their Future Therapeutic Potential. Pharm. Rev. 67, 821–870 (2015).
Wheeler, D. B., Randall, A. & Tsien, R. W. Roles of N-type and Q-type Ca2+ channels in supporting hippocampal synaptic transmission. Science 264, 107–111 (1994).
McGivern, J. G. Targeting N-type and T-type calcium channels for the treatment of pain. Drug Disco. Today 11, 245–253 (2006).
Striessnig, J., Ortner, N. J. & Pinggera, A. Pharmacology of L-type Calcium Channels: Novel Drugs for Old Targets? Curr. Mol. Pharm. 8, 110–122 (2015).
Wu, J. et al. Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350, aad2395 (2015).
Wu, J. et al. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 Å resolution. Nature 537, 191–196 (2016).
Zhao, Y. et al. Cryo-EM structures of apo and antagonist-bound human Cav3.1. Nature 576, 492–497 (2019).
Gao, S., Yao, X. & Yan, N. Structure of human Ca(v)2.2 channel blocked by the painkiller ziconotide. Nature 596, 143–147 (2021).
Yao, X., Gao, S. & Yan, N. Structural basis for pore blockade of human voltage-gated calcium channel Ca(v)1.3 by motion sickness drug cinnarizine. Cell Res. (2022).
Ertel, E. A. et al. Nomenclature of voltage-gated calcium channels. Neuron 25, 533–5 (2000).
Long, S. B., Campbell, E. B. & Mackinnon, R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005).
Tsien, R. W., Hess, P., McCleskey, E. W. & Rosenberg, R. L. Calcium channels: mechanisms of selectivity, permeation, and block. Annu Rev. Biophys. Biophys. Chem. 16, 265–90 (1987).
Corry, B., Allen, T. W., Kuyucak, S. & Chung, S. H. Mechanisms of permeation and selectivity in calcium channels. Biophys. J. 80, 195–214 (2001).
Hering, S. et al. Calcium channel gating. Pflug. Arch. 470, 1291–1309 (2018).
Tombola, F., Pathak, M. M. & Isacoff, E. Y. How does voltage open an ion channel? Annu Rev. Cell Dev. Biol. 22, 23–52 (2006).
Dong, Y. et al. Closed-state inactivation and pore-blocker modulation mechanisms of human Ca(V)2.2. Cell Rep. 37, 109931 (2021).
Parajuli, L. K. et al. Quantitative regional and ultrastructural localization of the Ca(v)2.3 subunit of R-type calcium channel in mouse brain. J. Neurosci. 32, 13555–67 (2012).
Schneider, T., Neumaier, F., Hescheler, J. & Alpdogan, S. Cav2.3 R-type calcium channels: from its discovery to pathogenic de novo CACNA1E variants: a historical perspective. Pflug. Arch. 472, 811–816 (2020).
Helbig, K. L. et al. De Novo Pathogenic Variants in CACNA1E Cause Developmental and Epileptic Encephalopathy with Contractures, Macrocephaly, and Dyskinesias. Am. J. Hum. Genet 103, 666–678 (2018).
Hainsworth, A. H., McNaughton, N. C., Pereverzev, A., Schneider, T. & Randall, A. D. Actions of sipatrigine, 202W92 and lamotrigine on R-type and T-type Ca2+ channel currents. Eur. J. Pharm. 467, 77–80 (2003).
Saegusa, H. et al. Altered pain responses in mice lacking alpha 1E subunit of the voltage-dependent Ca2+ channel. Proc. Natl Acad. Sci. USA 97, 6132–6137 (2000).
Benkert, J. et al. Cav2.3 channels contribute to dopaminergic neuron loss in a model of Parkinson’s disease. Nat. Commun. 10, 5094 (2019).
Matthews, E. A., Bee, L. A., Stephens, G. J. & Dickenson, A. H. The Cav2.3 calcium channel antagonist SNX-482 reduces dorsal horn neuronal responses in a rat model of chronic neuropathic pain. Eur. J. Neurosci. 25, 3561–3569 (2007).
Ellinor, P. T. et al. Functional expression of a rapidly inactivating neuronal calcium channel. Nature 363, 455–458 (1993).
Randall, A. & Tsien, R. W. Pharmacological dissection of multiple types of Ca2+ channel currents in rat cerebellar granule neurons. J. Neurosci. 15, 2995–3012 (1995).
Zhang, J. F. et al. Distinctive pharmacology and kinetics of cloned neuronal Ca2+ channels and their possible counterparts in mammalian CNS neurons. Neuropharmacology 32, 1075–1088 (1993).
Bourinet, E. et al. Interaction of SNX482 with domains III and IV inhibits activation gating of alpha(1E) (Ca(V)2.3) calcium channels. Biophys. J. 81, 79–88 (2001).
Huang, G. et al. High-resolution structures of human Nav1.7 reveal gating modulation through alpha-pi helical transition of S6IV. Cell Rep. 39, 110735 (2022).
Punjani, A. & Fleet, D. J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Zhao, Y. et al. Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca(2+) Channel. Cell 177, 1495–1506.e12 (2019).
Gao, S. & Yan, N. Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca(v) 1.1 by Dihydropyridine Compounds*. Angew. Chem. Int Ed. Engl. 60, 3131–3137 (2021).
Gregg, R. G. et al. Absence of the beta subunit (cchb1) of the skeletal muscle dihydropyridine receptor alters expression of the alpha 1 subunit and eliminates excitation-contraction coupling. Proc. Natl Acad. Sci. USA 93, 13961–13966 (1996).
Cheng, W., Altafaj, X., Ronjat, M. & Coronado, R. Interaction between the dihydropyridine receptor Ca2+ channel beta-subunit and ryanodine receptor type 1 strengthens excitation-contraction coupling. Proc. Natl Acad. Sci. USA 102, 19225–19230 (2005).
Schredelseker, J. et al. The beta 1a subunit is essential for the assembly of dihydropyridine-receptor arrays in skeletal muscle. Proc. Natl Acad. Sci. USA 102, 17219–17224 (2005).
Wormuth, C. et al. Review: Ca(v)2.3 R-type Voltage-Gated Ca(2+) Channels - Functional Implications in Convulsive and Non-convulsive Seizure Activity. Open Neurol. J. 10, 99–126 (2016).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7 (2018).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr 66, 213–221 (2010).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
DeLano, W. L. The PyMOL Molecular Graphics System, http://www.pymol.org.
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Hutchins, P. D., Russell, J. D. & Coon, J. J. LipiDex: An Integrated Software Package for High-Confidence Lipid Identification. Cell Syst. 6, 621–625.e5 (2018).
Ogiso, H. & Taguchi, R. Reversed-phase LC/MS method for polyphosphoinositide analyses: changes in molecular species levels during epidermal growth factor activation in A431 cells. Anal. Chem. 80, 9226–9232 (2008).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph 14, 354–360 (1996).
Acknowledgements
We thank the cryo-EM facility at Princeton Imaging and Analysis Center which is partially supported by the Princeton Center for Complex Materials (PCCM) and the NSF-MRSEC program (MRSEC; DMR-2011750). This work is supported by NIH grant (5R01GM130762 and 7R01GM057440-19, N.Y.; 1R01DK125404, Y.Y.), and Wellcome Trust grant (221795/Z/20/Z, D.W. and C.V.R).
Author information
Authors and Affiliations
Contributions
N.Y. conceived the project. X.Y., S.G., and N.Y. designed all experiments. X.Y. and S.G. conducted cryo-EM studies. Y.Y. supervised the electrophysiology study and Y.W. and Z.W. conducted the experiments and analyzed the data. C.V.R. supervised the mass spectrometry study and D.W. performed the experiments. X.F. performed 3D variability analysis. J.H. helped western blotting during manuscript revision. A.M., S.Ga. and M.H. helped manuscript preparation. All authors contributed to data analysis. N.Y. and X.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Doreen Matthies, Richard Tsien and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yao, X., Wang, Y., Wang, Z. et al. Structures of the R-type human Cav2.3 channel reveal conformational crosstalk of the intracellular segments. Nat Commun 13, 7358 (2022). https://doi.org/10.1038/s41467-022-35026-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-35026-6
This article is cited by
-
Structural basis for different ω-agatoxin IVA sensitivities of the P-type and Q-type Cav2.1 channels
Cell Research (2024)
-
Structural basis for human Cav3.2 inhibition by selective antagonists
Cell Research (2024)
-
Molecular insights into the gating mechanisms of voltage-gated calcium channel CaV2.3
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.