Abstract
Biodegradation of aromatic and heterocyclic compounds requires an oxidative ring cleavage enzymatic step. Extensive biochemical research has yielded mechanistic insights about catabolism of aromatic substrates; yet much less is known about the reaction mechanisms underlying the cleavage of heterocyclic compounds such as pyridine-ring-containing ones like 2,5-hydroxy-pyridine (DHP). 2,5-Dihydroxypyridine dioxygenase (NicX) from Pseudomonas putida KT2440 uses a mononuclear nonheme Fe(II) to catalyze the oxidative pyridine ring cleavage reaction by transforming DHP into N-formylmaleamic acid (NFM). Herein, we report a crystal structure for the resting form of NicX, as well as a complex structure wherein DHP and NFM are trapped in different subunits. The resting state structure displays an octahedral coordination for Fe(II) with two histidine residues (His265 and His318), a serine residue (Ser302), a carboxylate ligand (Asp320), and two water molecules. DHP does not bind as a ligand to Fe(II), yet its interactions with Leu104 and His105 function to guide and stabilize the substrate to the appropriate position to initiate the reaction. Additionally, combined structural and computational analyses lend support to an apical dioxygen catalytic mechanism. Our study thus deepens understanding of non-heme Fe(II) dioxygenases.
Similar content being viewed by others
Introduction
Pyridine rings are primary components of pyridoxyl derivatives, natural plant alkaloids, and coenzymes. These compounds are more soluble in water, meaning they can spread into groundwater, which are hazardous to the health of humans and other organisms1,2. A pyridine ring opening reaction step is a common feature of most chemical and enzyme-based degradation processes for such pollutants, yet relatively little is known about such reactions, highlighting that gaining a clearer understanding of pyridine ring opening should enable development of management technologies.
2,5-Hydroxy-pyridine (DHP), a potential carcinogen, is a metabolic intermediate in the catabolism of many pyridine derivatives3,4,5,6,7, which showed to cause DNA strand scission8. DHP is transformed to N-formylmaleamic acid (NFM) by a 2,5-DHP dioxygenase, an enzyme known as NicX from Pseudomonas putida KT2440 or Hpo from P. putida S165,9. A previous biochemical study showed that this enzyme is a mononuclear non-heme iron oxygenase9.
The superfamily of non-heme iron(II) enzymes catalyzes a wide range of oxidative transformations, ranging from the cis-dihydroxylation of arenes to the biosynthesis of antibiotics such as isopenicillin and fosfomycin10,11,12,13. These enzymes can be classified into several different groups based on their structural characteristics, reactivity, and specific requirements for catalysis, among them: (I) Extradiol cleaving catechol dioxygenases, (II) Rieske oxygenases, (III) Alpha-ketoglutarate dependent enzymes, (IV) Cysteine dioxygenases, and (V) Pterin-dependent hydroxylases13,14.
A phylogenetic analysis of non-heme Fe(II) dioxygenases indicted that NicX is a member of a subclass of the non-heme iron dependent oxygenases (Fig. 1)7. Unlike other known non-heme Fe(II) enzymes, NicX catalyzes a pyridine ring-cleavage7,15. Notably, many ring-cleavage dioxygenases have been reported, including 2,3-HPCD from Brevibacterium fuscum16, BphC from Pseudomonas sp. KKS10217, PcpA from Sphingobium chlorophenolicum18,19, and PnpCD from Pseudomonas sp. WBC-320. The substrates for all of these enzymes contain aromatic rings. NicX has strong specificity for its DHP substrate; it cannot catalyze ring opening for pyridine-ring containing phenols, hydroquinones, or catechols21. The typical structural motif consists of a mononuclear iron(II) metal center that is coordinated by two histidine residues and one carboxylate ligand from either a glutamate or an aspartate residue. This structural motif has been coined the “2-His-1-carboxylate facial triad”13. In addition, a His/His/His triad coordinated Fe(II) has been found in cysteine dioxygenases and gentisate dioxygenase22,23,24. Interestingly, NicX Ser302 coordinates the iron(II) ion; a similar metal ion-interacting serine residue has been reported for a dialkylglycine decarboxylase, Cu+-ATPases and for transcriptional activators like CueR and GolS25,26,27,28. Although NicX’s DHP dioxygenase activity was enzymologically characterized in the 1970s, its structure has remained unsolved.
In this study, we present kinetics, mutational, and structural studies of NicX and clarify how NicX selectively recognizes DHP. We solved a resting homo-hexameric NicX structure as well as a NicX–DHP–NFM complex structure. We found that four residues of NicX (His265, Ser302, His318, Asp320) coordinate the iron(II) ion. We also found that Leu104 and His105 adapt different conformations in DHP-bound monomers vs. NFM-bound monomers. In addition, molecular modeling, molecular dynamics simulations and quantum mechanics calculations were combined with the crystallographic 3D structural data to propose the plausible catalytic mechanisms of NicX. Our study of NicX deepens understanding of non-heme Fe(II) dioxygenases.
Results
Structural determination and overall structure
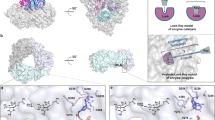
Seeking to better understand how NicX selectively recognizes its substrate DHP, we adopted a selenomethionine (SeMet) phasing strategy in which we initially determined the crystal structure of a SeMet-substituted resting NicX to a resolution of 2.28 Å using the single-wavelength anomalous dispersion (SAD) method (Table 1). Subsequently, the crystal structures of NicX and complex NicX–DHP–NFM were solved using coordinates of SeMet-NicX (Table 1). There are six molecules in an asymmetric unit in all the structures, consistent with gel filtration chromatography results revealing that NicX occurs as a homohexamer in solution (Fig. 2a)29.
a Overall structure of NicX. Ribbon plot representation of the NicX hexamer. b Overall structure of a protomer of NicX. The α-helices, β-sheets, and loops are in red, yellow, and green, respectively. Secondary structure elements of NicX are labeled. c Coordination sites of NicX-Fe(II). The 2FO-FC electron density map is contoured at 1σ, colored in blue; water 1 molecule is opposite Asp320, while water 2 molecule is opposite Ser302; green, red, blue, and purple represent carbon, oxygen, nitrogen, and Fe atoms, respectively. d The 14 Å-deep substrate-binding pocket on the NicX surface, two channels are observed. e Two channels are separated by residues His105 and Glu308, observed in resting or NFM complex structure. f In the DHP-bound subunits, channel II was blocked by the Leu104 and His105 residues.
The N-terminus of each NicX subunit has a 150-residue domain comprising 12 α helices and 19 β-strands (residues 2–151): this domain mainly consists of continuous αβ motifs. Each subunit also has a 199-residue C-terminal domain (residues 152–350), which contains two antiparallel β-sheets (β5-β6-β10-β13-β14-β18-β19/β7-β9-β11-β12-β15-β16-β17) (Fig. 2b).
Structure of the NicX–DHP–NFM complex
We examined the aforementioned NicX–DHP–NFM complex structure and found that only four subunits (C, D, E, and F) of the six constituent molecules in this structure bind with substrate DHP; the other subunits (A and B) contain the product NFM. This phenomenon of heterogeneous monomer binding patterns for a multi-homomeric enzyme is not surprising, with similar reports for other metal iron-dependent dioxygenases30,31,32,33. In the NicX–DHP–NFM complex, each subunit contains a fully occupied Fe(II), each of which coordinates with six ligands: four residues and two waters (Fig. 2c). Notably, site where water 2 is positioned exhibits elongated density in our models for one of the DHP-bound subunits (subunit D). This density was modeled as a water molecule, because the density for the possible oxygen is not clear while the previously reported trend that non-heme Fe(II) enzymes typically requires the presence of a substrate for oxygen binding to occur34,35.
Strikingly, the arrangement of the 14 Å-deep pockets in the NicX surface (Fig. 2d) changes based on the binding activity of a given subunit. That is, NFM-bound subunits appear the same as the resting state: there are apparently two channels at the enzyme surface near the active site, which are separated by residues His105 and Glu308, in close proximity to Leu104 (“open” state; Fig. 2e). However, only one of two channels occurs in the DHP-bound subunits (henceforth “channel I”, “close” state; Fig. 2f).
Characteristics of the Fe (II) coordination in NicX
There is a conspicuous, deep pocket (~14 Å in depth) on the surface of NicX that harbors its catalytic active site (Fig. 2d). Here, a ferrous ion is held in place via coordination involving four residues (His265, Ser302, His318,and Asp320) and two waters. Interestingly, the coordinating ligand Ser302 has not been previously reported in studies of other ferrous ion-dependent dioxygenases (Fig. 2c). To verify that these four residues directly participate in the iron coordination (rather than crystal packing), we conducted alanine screening mutation analysis. We observed a complete loss of activity for the H265A, S302A, H318A, and D320A mutants in enzymatic assays (Supplementary Table 1). Moreover, ICP-MS analysis revealed iron signals for the wild type enzyme but no such signals for any of these four mutant variants, and the circular dichroism analysis showed that the secondary structures of the mutation variants were not changed (Supplementary Table 2, Supplementary Table 3, and Supplementary Fig. 1). Collectively, these results verify the direct participation of these four residues in iron coordination with the C-terminal domain of NicX. It should be noted that although crystals were soaked in the buffer contain Fe2+, we were not able to experimentally determine the oxidation state of the iron in the crystal structures.
Substrate and product binding in the complex structure
In DHP-bound subunits of the complex, two hydrogen bonds are formed between Glu177 and the N1 and the O1 of DHP (2.5 and 3.0 Å), one hydrogen bond is formed between His189 and O1 of DHP (2.6 Å) (Fig. 3a). Thus, these hydrogen bond interactions may have an indispensable effect on the proper pre-catalytic positioning of the substrate. We explored the functional relevance of Glu177 and His189 with alanine mutant variants of these residues (as well as a NicXE177A&H189A mutant) which we tested with in vitro enzymatic assays with purified enzymes. Wild type NicX was purified to more than 95% homogeneity after recombinant expression in E. coli cells, and exhibited apparent Km and Vmax values for conversion of DHP to NFM of 94.9 ± 3.84 μM and 58.62 ± 0.95 U ∙ mg−1 (Supplementary Table 1), respectively. The purified NicXE177A&H189A variant completely lost its catalytic activity for DHP. The NicXE177A and NicXH189A variants enormously reduced activities (reduced to only 2.7 and 6.9% of the wild type), and exhibited 2.7-fold and 2.2-fold higher Km values compared with the wild type, respectively.
a Active site of NicX with bound DHP. The carbon atoms of DHP are in yellow. Green, red, blue, and purple represent carbon, oxygen, nitrogen, and Fe atoms, respectively. b Active site of NicX with bound NFM. The carbon atoms of NFM are in yellow. Cyan, red, blue, and purple represent carbon, oxygen, nitrogen, and Fe atoms, respectively. c A stereoview of the DHP bound subunit (green) and NFM bound subunit (cyan), the superimposition was done on the whole subunit. Iron and solvent molecules are shown as purple and red spheres, respectively. DHP/NFM interacts with residues were indicated as black dotted line, the coordination bonds of ferrous ion are shown as solid lines in purple, distances are given in angstroms.
It is intriguing that a NicXR293A variant completely lost activity for DHP (Supplementary Table 1). Observation in structure that the side chain of Arg293 is positioned about 4.3 Å distant from the center of DHP pyridine ring, and this residue does not apparently change its position or orientation in the resting or NFM-bound subunits (Fig. 3a). Moreover, Arg293 forms two salt bridge interactions with the side chain carboxyl group of the confirmed Fe-binding residue Asp320 (2.7 and 3.5 Å) (Fig. 3a). Pursuing this, we conducted ICP-MS assays and found that the NicXR293A variant lost the ability to bind a ferrous ion (Supplementary Table 2). This result indicates that the role of Arg293 may be primarily steric, apparently functioning to position the Asp320 correctly for ligation.
Analysis of the ring-open product NFM in the active sites of the A and B subunits of the complex structure revealed that the pyridine ring of DHP has been cleaved between the C5DHP and C6DHP carbons to form the product NFM (Fig. 3b). The carboxide derived from the C2DHP carbon of the product interacts particularly strongly with residues Glu177 (2.8 Å) and His189 (2.9 Å) (Fig. 3b), so that the product stably binds to the enzyme, additionally supported by NFM’s cis-carbon double bond7. As mentioned above, Fig. 3b provides direct evidence of ring fissure.
A conformational change of Leu104 and His105
A careful examination of structure shows that there is a conformational change for the Leu104 and His105 in the substrate-bound vs. both the resting structures and the product-bound subunits (Fig. 3c, Supplementary Movie 1). It is a surprise to find that a hydrophobic path that goes straight to the active center of ferrous ion when channel II is closed in the E subunit (Supplementary Fig. 2). However, this hydrophobic path is blocked by Leu104 in resting state or product NFM binding state, suggesting that a hydrophobic path appears induced by the conformational change of Leu104–His105 (Supplementary Fig. 2). Leu104 seems to interact with DHP through the side chain pyridine ring (3.9 Å) via both CH-π36 and van der Waals interactions (Fig. 3a). His105 forms a hydrogen bond with the substrate DHP (3.1 Å) (Fig. 3a). We performed in vitro assays with mutant variants to confirm functional contributions from these residues in NicX’s enzymatic activity. The NicXH105A variant completely lost enzyme activity, and the NicXL104A variant showed a dramatically decreased activity (down to only 3% of the wild type) and exhibited a 1.2-fold higher Km value compared with the wild type. Notably, the NicXE308A variant lost about half of its activity, and had a Km value very similar to wild type NicX, results suggesting that perhaps Glu308 does not participate directly in the recognition of DHP, but rather affects enzymatic activity via an interaction with His105.
Based on structural analysis, it could be seen from the NFM bound subunits that Cys76 is close to Leu104 at a distance of 4.9 Å, away from His105 at a distance of 7.7 Å (Fig. 4a, b). In contrast, in DHP bound subunits, Cys76 is close to His105 at a distance of 4.1 Å, away from Leu104 at a distance of 7.7 Å. This means that the conformational changes of Leu104–His105 may be influenced by Cys76 (Fig. 4a, b). It seems that the enzyme activity could also be influenced by Val175 in channel I but far away from active sites. To verify the above idea, Val175 in channel I and Cys76 in channel II were selected for mutation (Fig. 4a, b). The NicXV175F variant activity reduced to only 21.8% of the wild type, but Km values did not change much compared with the wild type, respectively (Supplementary Table 1). In contrast, the point mutations of C76Q or C76E completely abolished the enzymatic activity of NicX; however, it was notable that the NicXC76A variant retained the capability of transforming DHP into NFM (Supplementary Table 1). From the result of the mutation experiments, it appears that channel II has a more significant effect on the enzyme activity. Thus, we speculate that the bulky residues (glutamine or glutamic acid) take up more space than cysteine, and affect the range of movement of Leu104 and His105, causing it to be unable to guide and stabilize the substrate to the appropriate position to initiate the reaction.
a In NFM bound subunits, Cys76 is close to Leu104 at a distance of 4.9 Å, away from His105 at a distance of 7.7 Å. b In DHP bound subunits, Cys76 is close to His105 at a distance of 4.1 Å, away from Leu104 at a distance of 7.7 Å. Yellow, red, blue, and orange represent carbon, oxygen, nitrogen, and sulfur atoms, respectively.
Structural and computational studies for the NicX mechanism
Although NicX is the subtype defining member for non-heme iron(II) ring-cleavage dioxygenases, its overall catalytic mechanism bears some similarities with other non-heme iron(II) dioxygenases. Based on the studies of other Fe(II)-dependent dioxygenases12,13,30,31,32,37, and the computational analysis on our structures, we proposed two possible equatorial and apical dioxygen catalytic mechanisms, and calculated energetics of these pathways (Fig. 5, Supplementary Fig. 3).
Pathway IA and IB denote that the nitrogen and oxygen atoms of DHP bind the metal ferrous center, respectively. In Pathway II, the substrate DHP does not directly chelate with the ferrous ion. The possibly one-electron transfer radical species are drawn in square brackets. The C6 is the most vulnerable position in DHP, with an f- value of 0.148 based on Fukui function analysis in the upper left corner (C2, 0.067; C3, 0.062; C4, 0.052; C5, 0.090; C6, 0.148).
If the dioxygen takes the opposite position of Asp320 (“equatorial position”), there are two possibilities for substrates to coordinate with metal ions: the hydroxyl group is at position 5 to chelate with Fe (II) (Pathway IA), or the N atom on the pyridine ring can coordinate with Fe (II) (Pathway IB). Subsequently, two one-electron transfers are needed to form the peroxide intermediate, which can be followed by a Criegee rearrangement to yield a 7-membered-ring lactone and a ring-opened product; either of these scenarios would result in a substrate-bound iron arrangement similar to the classic extradiol catechol dioxygenases (Pathway IA & IB). Such a position would suggest a trans-effect for Asp320 in promoting the subsequent O-O cleavage reaction. However, this situation would require the DHP substrate to drop from the hydrogen-bonding network comprising His105–Glu177–His189, which would cause considerable destabilization of the reacting substrate–enzyme complex (Pathway IA & IB) (Supplementary Fig. 4a, b). Alternatively, the dioxygen molecule could be positioned opposite to Ser302 (“apical position”), in which case the C5 and C6 atoms of DHP would be adjacent to the dioxygen molecule. The apical arrangement would be similar to a P450-like arrangement, and Ser302 could exert a cystine-like catalytic role (Pathway II) (Supplementary Fig. 4c). More interestingly, the pyridinyl N-H could also plausibly involve in the proton transfer to activate the O-O cleavage in Pathway II, equivalent to imidazolyl N-H of the histidine residue in extradiol dioxygenases. Further investigation on the roles of Arg293–Asp320–DHPN-H is needed to illustrate the details of proton transfer for the O-O cleavage in the ring-open process. In a preliminary study of the conversion of the quintet peroxide intermediate to a 7-membered-ring lactone, both concerted, and stepwise mechanisms were examined (Pathway II, Supplementary Fig. 5). These calculations indicated a preference for a stepwise O-O cleavage; and low-lying septet state might promote spin-flip during the breaking and formation of multiple bonds (Supplementary Fig. 5).
Discussion
Multiple crystal structures of dioxygenases that can catalyze the ring-opening cleavage of aromatic compounds have been reported30,31,32, but we are unaware of any reported crystal structures for an enzyme capable of catalyzing cleavage of a pyridine ring. It is notable that a phylogenetic analysis of non-heme Fe(II) dioxygenases indicated that NicX and Hpo comprise a subclass of non-heme iron dependent oxygenases (Fig. 1). Despite extensive efforts, we were unable to obtain a structure for Hpo, and structural comparison using the DALI server failed to identify any obvious homologs of full-length NicX.
Another notable observation from our study that a conformational change in the Leu104 and His105 residues induce a major change in channel II; this change creates a hydrophobic path that goes straight to the active ferrous ion. Based on in vitro assays, we confirmed that His105 has a dramatic effect on enzyme activity. Previous studies have proposed that His residues could facilitate alkylperoxo intermediate breakdown, potentially via protonation to form a gem-diol intermediate38. We therefore examined whether His105 exerts this type of effect in NicX’s mechanism of action. However, we found that when His105 was mutated into aprotic amino acids (e.g., our NicXH105M and NicXH105F), these variants retained their catalytic activity for producing NFM from DHP (Supplementary Table 1). We interpret these results to rule out the participation of His105 in proton transfer during NicX catalysis. Our analyzes support speculation about two possible functions of the conformational change for Leu104–His105. The first is the aforementioned creation of a hydrophobic path that could greatly facility the direct delivery of a dioxygen molecule to the iron center (Supplementary Fig. 2). The second potential function of the conformational change could be to guide and assist to bind the DHP substrate molecule to a pre-reaction position, besides Glu177–His189; this may facilitate activation of the dioxygen and promote reaction initiation. Our site-directed mutagenesis results show that NicXC76Q and NicXC76E variants are inactivated provides further (albeit indirect) evidence of the contribution of Leu104 and His105 in orienting the substrate DHP.
It is also fascinating to find that the NicX ferrous center coordinates with only four residues (His265, Ser302, His318, and Asp320). Ser has been reported as a ligand for proteins including dialkylglycine decarboxylase, Cu+-ATPases, and transcriptional activators25,26,27,28; however, it has not been reported in non-heme dioxygenases, suggesting that NicX has a previously unknown fold architecture and active site environment for non-heme iron (II) dioxygenases. We examined site-directed mutant variants of NicX and confirmed the indispensability of these four residues for Fe(II) coordinating by using enzymatic assays, ICP-MS assays, and circular dichroism analyzes.
It is intriguing that NicX has not been reported to have activities for substrates other than DHP. Our study indicates that this substrate specificity is strongly impacted by NicX’s substrate-binding sites, including His105, His189, and Glu177, each of which is involved in the unique hydrogen bonding network. The O1 atom of DHP interacts with the residues of Glu177 and His189, and the N atom of DHP affects the electron distribution over the pyridine ring. Given that N atoms are more electronegative than carbon atoms, these factors apparently contribute to controlling the ionic configuration of the substrate DHP; thus the N atom of DHP is placed adjacent to the Glu177 side chain carboxyl group, rather than some other orientation. We consider the different catalytic pathways of NicX, wherein the dioxygen occupies the equatorial position (Pathway I) or apical position (Pathway II); these pathways are ultimately downhill energetically and cannot be distinguished based on energetics alone. However, the specific substrate orientation in the co-crystal structure motivated us to reconsider the possibility of Pathway II. First of all, the arrangement in Pathway II would allow maintenance of the specific hydrogen bonding network for holding the DHP substrate in place; in contrast, the DHP molecule would have to undergo a “big flip” and shift downwards by ~4 Å to bind the metal and contact the equatorial dioxygen in Pathway I. Importantly, this would require disturbing the specific hydrogen bond network observed in the crystal structure. Alternatively, in Pathway II the bridging peroxo can undergo a ~0.5 Å shift of the substrate and a ~20° rotation of the substrate, a scenario that would retain the intermediate in the hydrogen bond network. Second, the first one-electron transfer of the DHP substrate seems to occur at the moiety of O-C = N based on the pKa (pKa1 = 8.56) and Fukui function (f- = 0.148) (Fig. 5), and the dioxygen molecule could occupy the vacancy between C6---Fe and mediate the ignition of the DHP-O2-Fe triad. Third, the consequent O-O cleavage could be promoted by neighboring proton donor candidates such as imidazolium of His105, neutral form of Asp320, aromatic N-H of DHP, and even guanidinium of Arg293 39,40. Moreover, a crystal structure-based CAVER analysis indicated that the dioxygen tunnels terminate at a position opposite to Ser302 rather than Asp320 (Supplementary Fig. 2). Thus, both structural analysis and preliminary computation lend support the apical hypothesis, but additional theoretical calculations will be required for further validation on these possible mechanisms.
In past studies, the mechanisms of homogentisate 1,2-dioxygenase (HGDO, PDB code 4AQ6 [https://doi.org/10.2210/pdb4AQ6/pdb]) and homoprotocatechuate 2,3-dioxygenase (HPCD, PDB code 2IGA [https://doi.org/10.2210/pdb5XRN/pdb]) have been investigated by trapping different reaction cycle intermediates in different subunits of a single crystal30,32. HPCD is a prototypical type I extradiol dioxygenase acting on a catechol-type substrate, while HGDO reacts with a benzene-type substrate containing hydroxyl group in the para position. Both enzymes share things in common: (i) both catalyze the oxidative aromatic ring cleavage; (ii) both have a mononuclear iron(II) metal center that is coordinated by two histidine residues and one carboxylate ligand; (iii) their substrates act as dentate ligands when bound to the active site Fe (II) ion; (iv) their mechanisms both feature an attack of a superoxo ligand on their substrates at a hydroxylated carbon. It should be emphasized that both our crystal structure data and computational studies highlight differences in the apparent reaction mechanism of NicX (Fig. 5) compared to HPCD and HGDO. Specifically, (i) NicX is able to catalytically crack a pyridine ring substrate; (ii) NicX has a mononuclear iron(II) metal center that is coordinated by two histidine residues, one carboxylate and a serine residue; (iii) DHP does not directly chelate ferrous ion; (iv) the reaction between superoxide and DHP proceeds by reaction at the C6 atom of DHP, not the OH-group carrying C5 atom. In light of these differences, it is not surprising that our crystal structures and computational studies indicate clear distinctions for the proposed reaction mechanism of NicX vs. the reaction mechanisms of HPCD and HGDO.
In summary, we here determined the crystal structure of a pyridine ring-cleavage enzyme. Our structural and computational studies of NicX from P. putida shed lights on the ring-cleavage mechanisms used by dioxygenases; our study deepens understanding about how non-home Fe(II) ring-cleavage dioxygenase family enzymes interact with their aromatic and heterocyclic substrates.
Methods
Chemical reagents
2,5-Dihydroxypyridine was purchased from Aladdin. SeMet was purchased from Acros Organics. Crystallization screens were obtained from Hampton Research. All other chemicals were obtained commercially.
Plasmid preparation, recombinant expression, and protein purification
The DNA fragment encoding full-length WT P. putida KT2440 NicX was cloned into the pET-24a (Novagen) vector between the NdeI HindIII sites using DNA primers 5′-agtcatatgccggtgagcaatgcacaa-3′ and 5′-tataagctttcgcgctcgcgactcct-3′ (bearing a sequence encoding 6 C-terminal His-tags) (Supplementary Table 4). All mutants were generated using a whole-plasmid PCR and DpnI digestion method, and the sequences of the constructs were verified via Sanger sequencing, primer pairs used for installing each mutation were shown in Supplementary Table 4. These plasmids were transformed into E. coli BL21(DE3) cells. Cells possessing plasmids for the wild type and the mutant variant of NicX were grown at 37 °C to an OD600 of 0.8, after which they were subjected to overnight induction at 16 °C with 0.4 mM isopropyl β-D-1-thiogalactopyranoside. Centrifugation for 15 minutes at 4,000×g was used to harvest the cells, with the resulting cell pellets resuspended in a binding buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 20 mM imidazole, 0.5 mM PMSF, 1 mM FeCl2, 10 mM β-Mercaptoethanol) and subsequently lysed using a cell homogenizer. We then centrifuged the cell lysate and purified the supernatant with Ni2+-NTA affinity chromatography (Qiagen) and Superdex200 gel filtration chromatography (GE healthcare). The gel filtration buffer was comprised of 200 mM NaCl, 20 mM Tris-HCl (pH 8.0), and 2 mM dithiothreitol (DTT). Subsequently, fractions with bands putative recombinant NicX proteins (~39 kDA, assessed via SDS-PAGE) were combined and concentrated to 25 mg ml−1, and were then flash-frozen in liquid nitrogen and stored at −80 °C.
Note that a SeMet-substituted NicX variant was expressed using the methionine-autotrophic E. coli strain B834(DE3) cultured in M9 minimal media, and was purified using the same procedure as for the native protein, except that 5 mM DTT was used in the Superdex200 gel filtration buffer. Purity was monitored for all protein preparations based on SDS-PAGE, and protein concentrations were determined using a NanoDrop2000 spectrophotometer.
Enzyme assay
Purified WT and NicX mutant variant proteins used in the in vitro enzymatic activity assay. Activity assay was performed at 25 °C, monitoring the absorbance at 320 nm using a UV-Vis 2550 spectrophotometer21. The reaction mixture contained 20 mM Tris-HCl (pH 8.0), 50 µM FeCl2, as well as the DHP substrate and the enzyme (concentrations of both depending on the nature of the particular assay) in a total volume of 800 µl. The enzyme was incubated with FeCl2 (1 mM) for 1 min, and the reaction was initiated via the addition of DHP. Activity was defined as the amount of enzyme that catalyzes the conversion of 1.0 µmol of DHP in 1 min. The assay was performed independently three times, and data are presented as means ± the standard deviations.
Crystallization, data collection, and structure determination
Crystals of NicX were grown from a 1:1(v/v) mixture of a NicX protein solution (25 mg ml−1), a reservoir solution (0.2 M Succinic acid (pH 7.0) and 20% (w/v) PEG 3350), using the hanging-drop vapor-diffusion method at 20 °C. The SeMet-NicX variant was crystallized in the reservoir solution containing 0.2 M Sodium formate (pH 7.0) and 20% (w/v) PEG 3350. The complexes were prepared by soaking NicX crystals in cryoprotectant buffer supplemented with 5 mM Fe2+ for about 5 min prior to soaking crystals in cryoprotectant buffer supplemented with 5 mM DHP and 20 mM sodium dithionite. After soaking for 30–60 min, crystals were rapidly transferred into mother liquor solutions containing 25% glycerol prior to cryocooling in liquid nitrogen.
Crystal diffraction data sets of the native and SeMet-NicX were collected at the BL17U1 and BL19U1 beamlines of the Shanghai Synchrotron Radiation Facility by using an ADSC Quantum 315r detector or a DECTRIS PILATUS3 6 M detector at a wavelength of 0.97918 Å at 100 K. All data were processed and scaled using the HKL3000 program41. The SAD phases were determined using the Autosol module of PHENIX42. After the model-building with Coot43 and refinement with REFMAC44. The structures of NicX and complex NicX–DHP–NFM were solved using coordinates of SeMet-NicX; the substrate/product molecules were placed in the model based on the Fourier difference map, and refined using the geometric restraints prepared using REEL in Phenix45. The figures were prepared using PyMOL. Crystallographic statistics are listed in Table 1. The resulting coordinates and structure factors have been deposited in the Protein Data Bank (Protein Data Bank codes: 7CNT、7CUP、7CN3).
Inductively coupled plasma mass spectrometry
An inductively coupled plasma mass spectrometer, iCAP Qc (Thermo Fisher scientific, USA), with KED (Kinetic energy discrimination) system was used for detection. Before the ICP-MS assays, the purified mutant protein was incubated with an appropriate amount of ferrous ion for at least 30 min (the molar ratio of protein to ferrous ion was 1:5) and again subjected to a Superdex 200 column. 200 µl of protein sample was added 0.5 ml HNO3 and let it sit for half an hour prior to heat the water bath for 90 min. Before use, sample volume was diluted to 5 ml by water. The blank sample with the same amount of acid was prepared with the same procedure.
Secondary analysis of structure via circular dichroism spectra
We used JASCO J-815 to obtain the circular dichroism spectra of NicX and its related proteins, at 20 °C, while the samples were 0.2 mg ml−1 in 20 mM NaH2PO4-Na2HPO4 buffer (pH 7.4). We measured from 200 to 280 nm, with a cell length of 1.0 mm. We used the software CDPro to analyze the secondary structure compositions of the proteins.
Dynamic pH titration
The dynamic pH titrations were used to assess the dissociation constants (pKa) of DHP, in aqueous solutions. An ionic strength (I) of 0.1 mol L−1 KCl was used to perform the titrations and a pH range of 4.0–12.0 was used to perform the measurements.
MD simulation
All MD simulations were performed by using the Amber software suite46. The initial protein structure used in the molecular dynamic simulation was constructed with the crystal structures (PDB: 7CUP、7CN3). The enzyme-substrate complexation was referred with the catalytic mechanisms of the Catechol dioxygenases and Rieske oxygenases13,14. The dioxygen tunnels toward the Fe center were analyzed with CAVER 3.0 program47. Both two water-coordination sites were tested for dioxygen displacement. In particular, the peroxide intermediate was docked into the active site based on the crystal structures with AutoDock 4 program48. Appropriate substrate conformations were selected for multiple MD simulations. Protonation states of titratable residues were assigned at pH 6.5 using the H + + web server49, and Fe/substrate-binding residues were visually inspected then. The HF/6-31 G*//B3LYP/6-311 G* method was used to generate the substrate molecule in Gaussian 09, while the antechamber program was used to fit the RESP charge (restrained electrostatic potential charge)50. The python-based Metal Center Parameter Builder was used to build the force field of the iron center active site51. To prepare the topology and assess files of the enzyme-substrate complex, we used a cubic TIP3P water box (10 Å thick) from the surface of the complex, while sodium counter ion was used to neutralize the whole system. The particle mesh Ewald (PME) method was used to calculate the long-range electrostatic interactions in the MD simulation, while the SHAKE algorithm was used to constrain the hydrogen-involving bond lengths. We performed two minimization sequences, which relaxed the molecules of the solvents as well as the whole system. The temperature of the system increased from 0 to 300 K during 100 ps, at a collision frequency of 2 ps−1 of Langevin dynamics. We calibrated the system for 50 ps and collected the trajectory with constant pressure and temperature (NPT). Finally, 25 ns production simulations without any restraint were performed under NPT conditions. An integration time step of 2 fs was utilized with structural snapshots being extracted every 1000 steps. The simulation trajectory was analyzed by the cpptraj in Amber tools18.
QM calculations
Geometrical snapshots from the enzyme-peroxide MD cluster were extracted as the pre-reaction states (PRS)52 for the O-O and C-C bond breaking and forming, and were further subject to geometry optimization in Gaussian 09 program53. The quantum mechanical cluster model consisted of side chains of active site residues (His105, Glu177, His189, His265, Ser302, His318,and Asp320) and the iron cation and peroxide intermediate, which added up to 99 atoms and bore one positive charge. The optimization process was carried out at the level of ωB97X-D functional and LANL2DZ (Fe) and 6-31 G(d) basis sets in aqueous solution with the steered molecular dynamics approach54. The triplet, quintet, and septet potential energy surfaces were scanned along the C-C and O-O reaction coordinates55. All stationary point structures were further optimized with the same level of theory. Vibrational frequency analyzes were performed to ensure local minima or first-order saddle points, and the free energies were calculated at 298 K (standard condition). In addition, the intrinsic reaction coordinates calculations were carried out to identify transition states and immediate reactants and products.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data supporting the findings of this work are available within the paper and its Supplementary Information files or from the corresponding authors on reasonable request. Protein Data Bank (PDB): The coordinates and the structure factor amplitudes for the Se-NicX, NicX, and NicX complexed with ligands were deposited in Protein Data Bank under accession codes 7CNT, 7CUP, 7CN3; and have been released. Source data are provided with this paper.
References
Kaiser, J. P., Feng, Y. & Bollag, J. M. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 60, 483–498 (1996).
Orpin, C. G., Knight, M. & Evans, W. C. The bacterial oxidation of picolinamide, a photolytic product of diquat. Biochem. J. 127, 819–831 (1972).
Shukla, O. P. & Kaul, S. M. Microbiological transformation of pyridine N-oxide and pyridine by Nocardia sp. Can. J. Microbiol. 32, 330–341 (1986).
Tang, H. Z. et al. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep. 2, 377 (2012).
Tang, H. Z. et al. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 9, e1003923 (2013).
Cain, R. B., Houghton, C. & Wright, K. A. Microbial metabolism of the pyridine ring: metabolism of 2- and 3-hydroxypyridines by the maleamate pathway in Achromobacter sp. Biochem. J. 140, 293–300 (1974).
Jimenez, J. I. et al. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc. Natl Acad. Sci. USA 105, 11329–11334 (2008).
Kim, S. G. & Novak, R. F. Role of P450IIE1 in the metabolism of 3-hydroxypyridine, a constituent of tobacco smoke: redox cycling and DNA strand scission by the metabolite 2,5-dihydroxypyridine. Cancer Res. 50, 5333–5339 (1990).
Behrman, E. J. & Stanier, R. Y. The bacterial oxidation of nicotinic acid. J. Biol. Chem. 228, 923–945 (1957).
Solomon, E. I., Decker, A. & Lehnert, N. Non-heme iron enzymes: contrasts to heme catalysis. Proc. Natl Acad. Sci. USA 100, 3589–3594 (2003).
Bugg, T. D. H. Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron 59, 7075–7101 (2003).
Wackett, L. P. Mechanism and applications of Rieske non-heme iron dioxygenases. Enzym. Microb. Technol. 31, 577–587 (2002).
Bruijnincx, P. C., Koten, G. V. & Klein Gebbink, R. J. Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies. Chem. Soc. Rev. 37, 2716–2744 (2008).
White, M. D. & Flashman, E. Catalytic strategies of the non-heme iron dependent oxygenases and their roles in plant biology. Curr. Opin. Chem. Biol. 31, 126–135 (2016).
Harayama, S., Kok, M. & Neidle, E. L. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46, 565–601 (1992).
Miller, M. A. & Lipscomb, J. D. Homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum. A dioxygenase with catalase activity. J. Biol. Chem. 271, 5524–5535 (1996).
Kimbara, K. et al. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171, 2740–2747 (1989).
Xun, L. Y., Bohuslavek, J. & Cai, M. Characterization of 2, 6-dichloro-p-hydroquinone 1, 2-dioxygenase (PcpA) of Sphingomonas chlorophenolica ATCC 39723. Biochem. Biophys. Res. Commun. 266, 322–325 (1999).
Ohtsubo, Y. et al. PcpA, which is involved in the degradation of pentachlorophenol in Sphingomonas chlorophenolica ATCC39723, is a novel type of ring-cleavage dioxygenase. FEBS Lett. 459, 395–398 (1999).
Zhang, J. J., Liu, H., Xiao, Y., Zhang, X. E. & Zhou, N. Y. Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J. Bacteriol. 191, 2703–2710 (2009).
Gauthier, J. J. & Rittenberg, S. C. The metabolism of nicotinic acid. I. Purification and properties of 2,5-dihydroxypyridine oxygenase from Pseudomonas putida N-9. J. Biol. Chem. 246, 3737–3742 (1971).
Joseph, C. A. & Maroney, M. J. Cysteine dioxygenase: structure and mechanism. Chem. Commun. 32, 3338–3349 (2007).
Adams, M. A., Singh, V. K., Keller, B. O. & Jia, Z. C. Structural and biochemical characterization of gentisate 1,2-dioxygenase from Escherichia coli O157:H7. Mol. Microbiol. 61, 1469–1484 (2006).
Banerjee, A. et al. A structural model for the iron-nitrosyl adduct of gentisate dioxygenase. Eur. J. Inorg. Chem. 44, 4797–4804 (2018).
Toney, M. D., Hohenester, E., Cowan, S. W. & Jansonius, J. N. Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites. Science 261, 756–759 (1993).
González-Guerrero, M., Eren, E., Rawat, S., Stemmler, T. L. & Argüello, J. M. Structure of the two transmembrane Cu+ transport sites of the Cu+ -ATPases. J. Biol. Chem. 283, 29753–29759 (2008).
Ibáñez, M. M., Checa, S. K. & Soncini, F. C. A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J. Bacteriol. 197, 1606–1613 (2015).
Hobman, J. L. MerR family transcription activators: similar designs, different specificities. Mol. Microbiol. 63, 1275–1278 (2007).
Jiménez, J. I., Acebrón, I., García, J. L., Díaz, E. & Mancheño, J. M. A preliminary crystallographic study of recombinant NicX, an Fe2+-dependent 2,5-dihydroxypyridine dioxygenase from Pseudomonas putida KT2440. Acta Crystallogr. F. 66, 549–553 (2010).
Kovaleva, E. G. & Lipscomb, J. D. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science 316, 453–457 (2007).
Knoot, C. J., Purpero, V. M. & Lipscomb, J. D. Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase. Proc. Natl Acad. Sci. USA 112, 388–393 (2015).
Jeoung, J. H., Bommer, M., Lin, T. Y. & Dobbeka, H. Visualizing the substrate-, superoxo-, alkylperoxo-, and product-bound states at the nonheme Fe(II) site of homogentisate dioxygenase. Proc. Natl Acad. Sci. USA 110, 12625–12630 (2013).
Wilmot, C. M. & Pearson, A. R. Cryocrystallography of metalloprotein reaction intermediates. Curr. Opin. Chem. Biol. 6, 202–207 (2002).
Zhang, Y., Colabroy, K. L., Begley, T. P. & Ealick, S. E. Structural studies on 3-hydroxyanthranilate-3,4-dioxygenase: the catalytic mechanism of a complex oxidation involved in NAD biosynthesis. Biochemistry 44, 7632–7643 (2005).
Colabroy, K. L. et al. The mechanism of inactivation of 3-hydroxyanthranilate-3,4-dioxygenase by 4-chloro-3-hydroxyanthranilate. Biochemistry 44, 7623–7631 (2005).
Nishio, M., Umezawa, Y., Fantini, J., Weiss, M. S. & Chakrabarti, P. The CH-π hydrogen bonds in biological macromolecules. Phys. Chem. Chem. Phys. 16, 12648–12683 (2014).
Wilmot, C. M. Biochemistry. An ancient and intimate partnership. Science 316, 379–380 (2007).
Groce, S. L. & Lipscomb, J. D. Aromatic ring cleavage by homoprotocatechuate 2,3-dioxygenase: role of His200 in the kinetics of interconversion of reaction cycle intermediates. Biochemistry 44, 7175–7188 (2005).
Keenholtz, R. A. et al. Arginine as a general acid catalyst in serine recombinase-mediated DNA cleavage. J. Biol. Chem. 288, 29206–29214 (2013).
Burke, J. R. et al. Bifunctional substrate activation via an arginine residue drives catalysis in chalcone isomerases. ACS Catal. 9, 8388–8396 (2019).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Winn, M. D., Murshudov, G. N. & Papiz, M. Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Case, D. A., et al. AMBER 2018 (Univeristy of California, 2018).
Jurcik, A. et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34, 3586–3588 (2018).
Morris, G. M. et al. Autodock4 and autodocktools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Anandakrishnan, R., Aguilar, B. & Onufriev, A. V. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541 (2012).
Wang, J. M., Cieplak, P. & Kollman, P. A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21, 1049–1074 (2000).
Li, P. F. & Merz, K. M. MCPB.py: a python based metal center parameter builder. J. Chem. Inf. Model. 56, 599–604 (2016).
Shi, T. et al. Theoretical studies on the catalytic mechanism and substrate diversity for macrocyclization of pikromycin thioesterase. ACS Catal. 8, 4323–4332 (2016).
Frisch, M. J. et al. Gaussian09 Revision C.01 (Gaussian Inc., 2009).
Deng, C., Lam, W. H. & Lin, Z. Y. Computational studies on rhodium (III) catalyzed C-H functionalization versus deoxygenation of quinoline N-oxides with diazo compounds. Organometallics 36, 650–656 (2017).
Christian, G. J., Neese, F. & Ye, S. Unravelling the molecular origin of the regiospecificity in extradiol catechol dioxygenases. Inorg. Chem. 55, 3853–3864 (2016).
Acknowledgements
This work was supported by the grant of Ministry of Science and Technology of the People’s Republic of China (2018YFA0901200), by the grant from Science and Technology Commission of Shanghai Municipality (17JC1403300), by the “Shuguang Program” (17SG09) supported by Shanghai Educational Development Foundation, by Shanghai Excellent Academic Leaders Program (20XD1421900) from Science and Technology Commission of Shanghai Municipality, and by the grants from National Natural Science Foundation of China (31422004, 31970041, and 21377085). We thank Dr. John H. Snyder, Prof. Jiahai Zhou, and Prof. Shuangjun Lin for assistance and discussion regarding this manuscript.
Author information
Authors and Affiliations
Contributions
H.T. and G.L. designed the experiments. G.L., F.H., and X.O. performed the experiments. G.L., H.T., and Y-L.Z. wrote the manuscript. G.L., Y-L.Z., H.T., and P.X. revised the manuscript. P.Z., G.L., and H.T. analyzed the data. H.T., Y-L.Z., and P.X. conceived the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, G., Zhao, YL., He, F. et al. Structure-guided insights into heterocyclic ring-cleavage catalysis of the non-heme Fe (II) dioxygenase NicX. Nat Commun 12, 1301 (2021). https://doi.org/10.1038/s41467-021-21567-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-21567-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.