Abstract
Neuronal nitric oxide synthase (nNOS) plays a crucial role in the maintenance of correct skeletal muscle function due, at least in part, to S-nitrosylation of specific protein targets. Similarly, we recently provided evidence for a muscular phenotype in mice lacking the denitrosylase S-nitrosoglutathione reductase (GSNOR). Here, we demonstrate that nNOS and GSNOR are concomitantly expressed during differentiation of C2C12. They colocalizes at the sarcolemma and co-immunoprecipitate in cells and in myofibers. We also provide evidence that GSNOR expression decreases in mouse models of muscular dystrophies and of muscle atrophy and wasting, i.e., aging and amyotrophic lateral sclerosis, suggesting a more general regulatory role of GSNOR in skeletal muscle homeostasis.
Similar content being viewed by others
Introduction
The role of nitric oxide (NO) in skeletal muscle homeostasis has been deeply investigated, and findings of the last decades support the hypothesis that it is involved in both muscle contraction and atrophy1,2,3,4. In rat models of denervation- and disuse-induced atrophy5, and in dystrophin-null (mdx) mouse models of genetic dystrophy2,6,7, it has been observed that the skeletal muscle specific form of neuronal NO synthase (nNOS) dislocates from the dystrophin glycoprotein complex (DGC) located at the sarcolemma8. This leads first to the loss of NO beneficial effects, mostly exerted via cGMP signaling (e.g., vasodilation and satellite cell proliferation)1,9, and second to nitration and hyper-S-nitrosylation of several proteins, including those involved in Ca2+ release (i.e., type 1 ryanodin receptor, RyR1)10, in stress response and apoptosis (e.g. NF-κB and FoxO3)5,11.
We previously demonstrated that mice lacking the denitrosylase S-nitrosoglutathione reductase (GSNOR) show muscular atrophy characterized by atrogenes expression, mitochondrial alteration, and apoptosis12, suggesting that defective denitrosylation affects skeletal muscle function. Interestingly, we also observed that two mouse models of genetic dystrophies, namely the mdx13 and α-sarcoglycan-deficient (α-SG−/−) mice14, showed excessive levels of S-nitrosylated proteins (PSNOs) similar to those detected in GSNOR-null (Gsnor−/−) mice, suggesting that aberrant S-nitrosylation is a hallmark of muscle wasting.
It has been reported that GSNOR and nNOS co-localize with type 2 ryanodine receptor (RyR2) along the T-tubular invaginations of cardiac myocytes, this being crucial for regulation of vascular tone and cardiac contractility15,16,17. These results strongly suggest that GSNOR and nNOS act in concert to dynamically regulate NO flux and convey it on specific targets.
Here we provide evidence that GSNOR and nNOS co-immunoprecipitate and co-localize, reasonably at the sarcolemma, and that GSNOR expression is required for correct skeletal muscle differentiation and homeostasis.
Results and discussions
S-nitrosylation increase and GSNOR reduction are events associated with muscular atrophy and aging
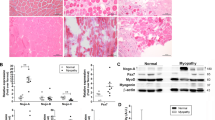
We previously reported that skeletal muscles from mdx and αSG−/− dystrophic mice show PSNOs increase that correlates with a decrease in GSNOR expression12. To give strength to this observation, we evaluated PSNOs and GSNOR levels in another in vivo model of progressive muscle atrophy. Particularly, we focused on mouse models of familial amyotrophic lateral sclerosis (fALS) expressing the G93A-SOD1 mutant18, either systemically or exclusively in the skeletal muscle (MLC-SOD1G93A)19. Biotin switch assays and western blot analyses of gastrocnemius indicate that, similarly to Gsnor−/− (KO) and mdx mice, both fALS models show an increase of PSNOs (Fig. 1a). These results inversely correlate with GSNOR levels (Fig. 1b, c), supporting the idea that decreased GSNOR expression and the resulting excessive S-nitrosylation are two signatures of atrophic muscle.
Total homogenates of gastrocnemius from 2-month-old wild-type (WT), Gsnor−/− (KO), mdx mice, and mouse models of fALS expressing the G93A-SOD1 mutant, either systemically (SOD1G93A), or exclusively in the skeletal muscle (MLC-SOD1G93A) were used for: a Biotin-switch assays of S-nitrosylated proteins (PSNOs) revealed, upon biotynilation, by incubation with horseradish peroxidase (HRP)-conjugated streptavidin. b Western blot analyses of GSNOR levels. Lactate dehydrogenase (LDH) was selected as a loading control both in (a) and (b). Results shown are representative of 3 that gave similar results. (c) Densitometry of GSNOR bands shown in (B), calculated by FiJi analysis software. Values shown are the means±SD of n = 3 different experiments normalized to LDH
Muscular atrophy is a condition usually associated with physio-pathological states related to disuse (e.g. aging), in which regeneration rate is decreased and skeletal muscle size and performance coherently reduced20,21. We also have recently demonstrated that GSNOR expression levels are reduced during aging. As a consequence, protein S-nitrosylation increases, this being a distinctive feature of aging in mammals22. Real-time qPCR and western blot analyses performed in skeletal muscle from 2-to-12 months old WT mice indicate that GSNOR mRNA and protein levels decrease with age also in this tissue (Fig. 2a–c). Coherently, PSNOs increase (Fig. 2d), suggesting that GSNOR hypo-expression is generally associated with a dysfunctional/aged skeletal muscle.
a Representative western blot of GSNOR levels in gastrocnemius homogenates from 2-, 6-, and 12-months-old WT mice. Homogenates obtained from 2-months-old Gsnor−/− (KO) muscle are shown as negative control of GSNOR expression. Lactate dehydrogenase (LDH) was selected as a loading control. b Densitometry of GSNOR bands shown in a, calculated by FiJi analysis software. Values shown are the means±SD of n = 3 different experiments normalized to LDH. c RT q-PCR analysis of GSNOR performed in gastrocnemius homogenates from 2- and 12-months-old WT mice. Results shown are the means±s.e.m. of n = 6 animals for each group. ***p < 0.001. d Representative biotin-switch assay of S-nitrosylated proteins (PSNOs) performed in total homogenates of gastrocnemius obtained from 2-months-old Gsnor−/− (KO), and from 2-, 6-, and 12-months-old WT mice (WT)
GSNOR and nNOS co-localize and co-immunoprecipitate in the skeletal muscle and in myoblasts
S-nitrosylation is a posttranslational modification whose extent depends on the balance between the rates of NO production and denitrosylation, with the latter reaction largely catalyzed by GSNOR23,24,25. In the skeletal muscle, nNOS is the main enzyme responsible for NO production, which predominantly shows a sarcolemmal distribution26. Based on previous results suggesting an interaction between GSNOR and nNOS in cardiomyocytes16,27, we hypothesized that, even in the skeletal muscle, GSNOR regulates S-nitrosylation extent by positioning in close contact with nNOS. Therefore, we first investigated the localization of GSNOR. Immunofluorescence analyses of tibialis anterior sections show that GSNOR localizes at the sarcolemma (Fig. 3a), where also Collagen III is located. We previously showed that GSNOR deficiency did not produce any alterations in sarcolemmal nNOS distribution12. This suggests that, notwithstanding the same localization, GSNOR does not apparently affect nNOS attachment to DGC and, in turn, subcellular localization of NO production.
a Representative fluorescence microscopy images of tibialis anterior sections from wild-type (WT) mice stained with anti-GSNOR and anti-collagen III (Col III) antibodies. b Immunoprecipitation (IP) of tibialis anterior homogenates from 2-months-old WT and Gsnor−/− (KO) mice performed with an anti-nNOS antibody and revealed by western blot (IB) for GSNOR and nNOS. Gsnor−/− (KO) muscles were selected as control. c Immunoprecipitation (IP), as described in b, of lysates from C2C12 myoblasts induced to differentiate upon 4 days of serum deprivation. d Representative fluorescence microscopy images of human specimens of rectus abdominis stained with anti-GSNOR and anti-nNOS antibodies, or e with anti-GSNOR and DAPI to highlight the nuclear/perinuclear localization
On the basis of this result, we performed co-immunoprecipitations in tibialis anterior lysates and in differentiating C2C12 mouse myoblasts. Results shown in Fig. 3b, c indicate, for the first time, that the two proteins were present in the same complex, suggesting that GSNOR – although not affecting nNOS localization – may still modulate NO effects on proteins located at the DGC. Interestingly, we were not able to observe any co-immunoprecptation in C2C12 cells before 4 days of differentiation, time at which the process of myogenesis is in an advanced state. This suggests that the presence of GSNOR and nNOS in the same complex might play a role in the late phases of differentiation. In support to these results, immunofluorescence analyses of human specimens of rectus abdominis confirmed that GSNOR co-localizes with nNOS (Fig. 3d) and, in line with previous reports28, also shows a (peri)nuclear distribution (Fig. 3e).
GSNOR contributes to muscle cell differentiation
To verify the hypothesis that GSNOR is directly involved in skeletal muscle differentiation and homeostasis, we investigated about the existence of an integrated regulation between GSNOR and nNOS. To this end, we evaluated their levels by western blot analysis in differentiating C2C12 mouse myoblasts. Figure 4a shows that nNOS and GSNOR increase time-dependently in parallel with syntrophin (used as marker of differentiation). Next, we downregulated GSNOR by short-hairpin RNA (shRNA) (Fig. 4b), and analyzed if this induced any alterations in C2C12 differentiation. Western blot analyses of myogenin and myosin heavy chain (MHC) – two proteins required respectively for commitment and differentiation of myogenic precursor cells29 – indicate that their expression is decreased upon GSNOR knocking-down (Fig. 4c). Of note, GSNOR-depleted (shGSNOR) C2C12 cells display a reduction in nNOS levels (Fig. 4d), exhibit a decreased number of myotubes and a lower fusion index with respect to the control (shScr) counterparts (Fig. 4e).
a Representative western blot analysis of GSNOR and nNOS in cell lysates from C2C12 upon serum deprivation-induced differentiation. Undifferentiated myoblasts (day 0); differentiating myotubes (day 1–4). Syntrophin and superoxide dismutase 1 (SOD1) were selected as differentiation and loading control, respectively. b, c Representative western blot analysis of nNOS, GSNOR (b) and myogenin and myosin heavy chain (MHC) (c) in non-differentiated shGSNOR and shScr C2C12 (day 0), or in the same cells after 4-days of serum deprivation-induced differentiation (day 4). SOD1 and LDH were selected as a loading control. Densitometry of each lane intensity is shown beneath the western blots. It was calculated by FiJi analysis software, normalized to LDH and arbitrarily set to 1.0 in shScr cells. d Immunofluorescence analysis of shGSNOR and shScr myoblasts (day 0), or 4-days differentiated myotubes (day 4). Nuclei were visualized upon staining with Hoechst 33342. Transfected cells were visualized taking advantage of the green-fluorescence of shRNAs. Superimposition (Merge) was used to calculate the fusion index, shown on the right. e Quantification of the data. Results are expressed as number of nuclei per fiber and represent the means±s.d. of n = 30 fibers (myotubes). ***p < 0.001
These results strongly argue for GSNOR playing a role in myogenesis, this being in perfect agreement with recent data30 and previous evidence showing that Gsnor−/− mice show a delayed muscle regeneration following injury12.
Conclusion
The role of NO and S-nitrosylation in skeletal muscle homeostasis has been exhaustively studied. However, the implication of GSNOR and denitrosylation remains controversial12,31. Here, we provide evidence that, in the same way of kinases and phosphatases (and/or ligases and de-ubiquitinylases), the denitrosylating enzyme GSNOR co-localizes with the source of NO, nNOS, to precisely control S-nitrosylation. Together with recent data indicating that, at least in E.coli, S-nitrosylation is an enzymatically driven process32, our results represents a further evidence that S-nitrosylation is a finely controlled posttranslational modification which is required, in the skeletal muscle, to maintain correct myofiber function and homeostasis. Such a fine regulation is lost when nNOS dislocates from sarcolemma, or – as here reported – when GSNOR is downregulated. Actually, besides excessive S-nitrosylation, other NO-mediated mechanisms concur to muscular atrophy, which we cannot exclude might also play a role in GSNOR-deficient systems, such as: (1) deactivation of cGMP signaling, which is extremely important to sustain vasodilation1,26 and stimulate satellite cells proliferation9; (2) deregulation of Ca2+ uptake/release for sarcoplasmic reticulum1,33; (3) impairment of mitochondrial biogenesis and metabolism34.
In line with data arguing for a pivotal role of GSNOR and S-nitrosylation in myoblast differentiation30 and muscle regeneration12,31, we also observed that GSNOR is involved in myogenesis. This is probably due to its recruitment in the same complex with nNOS, which becomes detectable at day 4 when other markers of differentiation are still expressed (Fig. 5). This evidence suggests that nNOS and GSNOR start to functionally interact close to the sarcolemma in the late phases of muscle differentiation. GSNOR is found decreased in aging and in genetic models of muscular atrophy, this allowing to speculate that this phenomenon contributes to skeletal muscle homeostasis by conveying NO signal on specific protein thiols located underneath the sarcolemma (Fig. 5). Linked to this, it has been demonstrated that mesenchymal stem cells from Gsnor−/− mice exhibit lower adipogenic versus higher osteogenic differentiation, due to an inhibitory S-nitrosylation at Cys139 in PPARγ35. However, Gsnor−/− mice are smaller than WT counterparts and exhibit bone loss due to an increased number of osteoclasts36. This suggests that selective S-nitrosylation underlies skeletal muscle homeostasis by means of at least two different mechanisms: (i) directly, as above mentioned, by affecting stem cell differentiation and tissue regeneration; (ii) indirectly, at systemic level, by interfering with the mechanic stimuli aimed at inducing an appropriate skeletal muscle implant into the bone.
Our data indicate that GSNOR and nNOS are co-expressed and reasonably part of the same complex (co-immunoprecipitate) during myogenesis. In this way, they sustain differentiation (left). Co-immunoprecipitation and co-localization analyses indicate that GSNOR and nNOS might be recruited in the same complex close to the sarcolemma. This, along with the correct activation of NO/cGMP signaling induced by sarcolemmatic nNOS, should allow the nitrosylation of specific targets (e.g. RyR1) and contribute to the correct physiology of the skeletal muscle (e.g., vasodilation and regeberation) (middle). During aging, or in dystrophic muscles, GSNOR levels are found to be reduced and, coherently, the extent of S-nitrosylated protein (PSNO) increased (right). GSNOR decrease is a hallmark of patho-physiological states associated with muscle wasting. However, we still do not know if this event is associated with nNOS translocation into the sarcoplasm, as reported in different pathological models of atrophy. Although reasonable, so far there is no evidence supporting this hypothesis
In agreement with these observations, two-month-old Gsnor−/− mice exhibit both osteopenia35 and sarcopenia12, which represent pathological states related to aging. We recently reported that GSNOR is physiologically silenced in mouse and human aging due to downregulation of the DNA demethylase, ten-eleven translocation protein 1 (Tet1)22. Remarkably, Tet1−/− mice are smaller than WT animals, resembling, in such an aspect, Gsnor−/− mice. This feature is associated with developmental skeletal muscle defects and deregulated expression of muscle contraction genes, which correlate with an augmented methylation status of the DNA37. Here, we have coherently provided evidence that in mouse models of muscular dystrophies, or physio-pathological states associated with skeletal muscle wasting (i.e. aging and ALS), GSNOR expression is decreased. These results argue for GSNOR being a molecular determinant of skeletal muscle homeostasis, with any dysregulations of its expression potentially affecting muscle healthy state.
Materials and methods
Animals
Mouse experiments were carried out in accordance with the European Community guidelines and with the approval of relevant National and local ethical committees. The Gsnor−/− mouse strain was generated by Prof. Stamler15,38, while the C57BL/6 wild-type were purchased from Charles River. mdx and α-SG−/− mice were from Jackson Laboratories. G93A-SOD1mice B6.Cg-Tg(SOD1 G93A)1Gur/J were purchased from The Jackson Laboratory and were kept on C57BL/6 J background. Mouse models of fALS expressing the mutant SOD1G93A selectively in skeletal muscle (MLC-SOD1G93A) were generated by Prof. Musarò19. Mice were housed in an environmentally controlled room (23 °C, 12 h light–dark cycle) and provided with food and water ad libitum.
Detection of PSNOs
Protein S-nitrosylation extent was evaluated by biotin-switch assay as previously described12,39. In brief, muscles were homogenized in HEN buffer (25 mM HEPES, 50 mM NaCl, 0.1 mM EDTA1% NP-40, protease inhibitors, pH 7.4). Free cysteine residues were blocked with S-methyl methanethiosulfonate (MMTS, Sigma) and protein pellets, collected upon precipitation in cold acetone, were re-suspended in HENS buffer (HEN buffer with 1% SDS) and let react with biotin-HPDP, with or without ascorbate. Biotinylated proteins were revealed using the Amersham ECL detection system after incubation with the HRP-conjugated streptavidin (Merck).
Immunoprecipitation assays
Immunoprecipitations were performed adding 1 μg of anti-nNOS antibody (Santa Cruz), or IgG, to 10 μl of prewashed Dynabeads®-protein G (Invitrogen). Five hundred microgram of whole tibialis anterior extracts, or 800 μg of C2C12 cell extracts, were incubated for 3 h with Dynabeads®-Ab complex and washed with 0.15 M NaCl, 10 mM HEPES pH 7.5. Immunoprecipitated proteins were detached from beads by boiling in sample buffer, separated by SDS-PAGE, transferred to nitrocellulose membranes (Amersham) and then incubated with anti-GSNOR or anti-nNOS antibody. IgG were used as negative control.
RT q-PCR
Gastrocnemius was homogenized in TRI-Reagent (Sigma) and RNA was extracted in accordance with the manufacturer’s protocol. cDNA was generated using the GoScript Reverse Transcription System (Promega). RT q-PCR was performed using the iTAQ Universal SYBR Green Supermix (Bio-Rad) on ViiA 7 Real-Time PCR System (Thermo-Fisher Scientific) and data were analyzed using the second derivative maximum method. All reactions were run as triplicates and normalized to the internal standard ribosomal protein L34. Primers used are the following:
GSNOR FW-tcacttcatggggactagca, RV-ccgagggatcgattttagca;
L34 FW-ggtgctcagaggcactcaggatg, RV- gtgctttcccaaccttcttggtgt.
Cell culture and transfection
C2C12 cells were grown in DMEM (Thermo-Fisher Scientific) supplemented with 10% FBS, 1000 U/mL penicillin-streptomycin at 37 °C in 5% CO2, or in 2% horse serum-containing DMEM to induce differentiation. When stated, 24 h after plating, C2C12 cells were transfected with short-hairpin RNAs against GSNOR designed in our laboratory and synthesized by Sigma.
Top strand:
5′-tgctgctcccactaccacactgacacgttttggccactgactgacgtgtcagtggtagtgggag-3′;
Bottom strand:
5′-cctgctcccactaccactgacacgtcagtcagtggccaaaacgtgtcagtgtggtagtgggagc-3′. The oligonucleotides were cloned in the pcDNA6.2-GW/EmGFP-miR vector (Thermo-Fisher Scientific) using the BLOCK-iT™ Pol II miR RNAi Expression Vector Kit with EmGFP (Thermo-Fisher Scientific) in according to manufacturer’s instructions.
Immunofluorescence
Human tissues: muscle biopsy specimens were derived from a previous study (https://doi.org/10.1038/srep30340). They were obtained from the M.G. Vannini Hospital in Rome (Italy), from patients who signed an informed consent, after clearance by the local ethical committee. Biopsy specimens were obtained during the initial phase of the operation from the rectus abdominis muscle of patients undergoing abdominal surgery for non-neoplastic reasons, and used as controls in the previous study. Reasons for abdominal surgery in controls were incisional hernia, cholelithiasis, benign prostatic hyperplasia, epigastric hernia and mesenteric cyst. Biopsy specimens were immediately frozen in liquid nitrogen and stored at –80 °C until analysis. Mouse tissues: tibialis anterior and gastrocnemius were embedded in O.C.T. (Bio-Optica) and flash-frozen in liquid nitrogen-cooled isopentane (VWR). All sections were cut to a thickness of 8 μm using a Leika cryostat; fixed in 4% paraformaldehyde; permeabilized in 0.2% Triton X-100/1% BSA (Sigma); blocked in 10% horse serum (Sigma); and incubated for 1 h with: anti-GSNOR (Sigma, 1:100) and anti-Collagen III (Sigma, 1:100). Afterwards, cryosections were incubated with labeled secondary antibodies (Thermo-Fisher Scientific) and examined by a Leica TCS-SP5 confocal microscopy. Fluorescence images were adjusted for brightness, contrast, and color balance using Fiji40. Cells: undifferentiated myoblasts (day 0) and differentiated myotubes (day 4) were fixed in 4% paraformaldehyde, stained with Hoechst 33342 (to visualize nuclei) and analyzed using an EVOS Floid Cell Imaging Station (Thermo-Fisher Scientific). Only cells/fibers expressing the GFP-tagged shRNAs were considered to quantify the fusion index, which has been evaluated by counting the number of nuclei in each fiber with Fiji analysis software40.
Western blotting
Samples from gastrocnemius were homogenized, and C2C12 cells were lysed in lysis buffer containing 0.15 M NaCl, 10 mM HEPES (pH 7.5). Antibodies used: anti-nNOS, anti-SOD1, anti-LDH, anti-α-syntrophyn, anti-myogenin, anti-MHC (Santa Cruz), and anti-GSNOR (Millipore). Immune-reactive bands were revealed by Chemidoc System (Bio-Rad) and quantified by densitometry using Fiji40.
Protein concentration was determined by the DC™ Protein Assay (Bio-Rad).
References
Stamler, J. S. & Meissner, G. Physiology of nitric oxide in skeletal muscle. Physiol. Rev. 81, 209–237 (2001).
Bellinger, A. M. et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat. Med. 15, 325–330 (2009).
Bellinger, A. M. et al. Remodeling of ryanodine receptor complex causes “leaky” channels: a molecular mechanism for decreased exercise capacity. Proc. Natl Acad. Sci. USA 105, 2198–2202 (2008).
Grozdanovic, Z. NO message from muscle. Microsc. Res. Tech. 55, 148–153 (2001).
Suzuki, N. et al. NO production results in suspension-induced muscle atrophy through dislocation of neuronal NOS. J. Clin. Invest. 117, 2468–2476 (2007).
Grozdanovic, Z., Gosztonyi, G. & Gossrau, R. Nitric oxide synthase I (NOS-I) is deficient in the sarcolemma of striated muscle fibers in patients with Duchenne muscular dystrophy, suggesting an association with dystrophin. Acta Histochem. 98, 61–69 (1996).
Brenman, J. E., Chao, D. S., Xia, H., Aldape, K. & Bredt, D. S. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell 82, 743–752 (1995).
Crosbie, R. H., Barresi, R. & Campbell, K. P. Loss of sarcolemma nNOS in sarcoglycan-deficient muscle. FASEB J. 16, 1786–1791 (2002).
Buono, R. et al. Nitric oxide sustains long-term skeletal muscle regeneration by regulating fate of satellite cells via signaling pathways requiring Vangl2 and cyclic GMP. Stem Cells 30, 197–209 (2012).
Andersson, D. C. et al. Leaky ryanodine receptors in β-sarcoglycan deficient mice: a potential common defect in muscular dystrophy. Skelet. Muscle 2, 9 (2012).
Inadomi, C. et al. Overexpression of glutaredoxin protects cardiomyocytes against nitric oxide-induced apoptosis with suppressing the S-nitrosylation of proteins and nuclear translocation of GAPDH. Biochem. Biophys. Res. Commun. 425, 656–661 (2012).
Montagna, C. et al. S-nitrosoglutathione reductase deficiency-induced S-nitrosylation results in neuromuscular dysfunction. Antioxid. Redox Signal. 21, 570–587 (2014).
Partridge, T. A. The mdx mouse model as a surrogate for Duchenne muscular dystrophy. FEBS J. 280, 4177–4186 (2013).
Hack, A. A. et al. Muscle degeneration without mechanical injury in sarcoglycan deficiency. Proc. Natl Acad. Sci. USA 96, 10723–10728 (1999).
Liu, L. et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628 (2004).
Beigi, F. et al. Dynamic denitrosylation via S-nitrosoglutathione reductase regulates cardiovascular function. Proc. Natl Acad. Sci. USA 109, 4314–4319 (2012).
Lima, B., Forrester, M. T., Hess, D. T. & Stamler, J. S. S-nitrosylation in cardiovascular signaling. Circ. Res. 106, 633–646 (2010).
Gurney, M. E. et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994).
Dobrowolny, G. et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 8, 425–436 (2008).
Domingues-Faria, C., Vasson, M. P., Goncalves-Mendes, N., Boirie, Y. & Walrand, S. Skeletal muscle regeneration and impact of aging and nutrition. Ageing Res. Rev. 26, 22–36 (2016).
Bodine, S. C. Disuse-induced muscle wasting. Int. J. Biochem. Cell Biol. 45, 2200–2208 (2013).
Rizza, S. et al. S-nitrosylation drives cell senescence and aging in mammals by controlling mitochondrial dynamics and mitophagy. Proc. Natl Acad. Sci. USA 10, E3388–E3397 (2018).
Rizza, S. & Filomeni, G. Chronicles of a reductase: biochemistry, genetics and physio-pathological role of GSNOR. Free Radic. Biol. Med. 110, 19–30 (2017).
Jensen, D. E., Belka, G. K. & Du Bois, G. C. S-nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem. J. 331, 659–668 (1998).
Hess, D. T. & Stamler, J. S. Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 287, 4411–4418 (2012).
Kobayashi, Y. M. et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature 456, 511–515 (2008).
Sips, P. Y. et al. Reduction of cardiomyocyte S-nitrosylation by S-nitrosoglutathione reductase protects against sepsis-induced myocardial depression. Am. J. Physiol. Heart Circ. Physiol. 304, H1134–H1146 (2013).
Iborra, F. et al. Immunocytochemical and biochemical demonstration of formaldehyde dehydrogenase (class III alcohol dehydrogenase) in the nucleus. J. Histochem. Cytochem. 40, 1865–1878 (1992).
Shimokawa, T., Kato, M., Ezaki, O. & Hashimoto, S. Transcriptional regulation of muscle-specific genes during myoblast differentiation. Biochem. Biophys. Res. Commun. 246, 287–292 (1998).
Yamashita, A. M. S. et al. Balance between S-nitrosylation and denitrosylation modulates myoblast proliferation independently of soluble guanylyl cyclase activation. Am. J. Physiol. Cell Physiol. 313, C11–C26 (2017).
Moon, Y. et al. GSNOR deficiency enhances in situ skeletal muscle strength, fatigue resistance, and RyR1 S-nitrosylation without impacting mitochondrial content and activity. Antioxid. Redox Signal. 26, 165–181 (2016).
Seth, D. A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol. Cell 69, 451–464.e6 (2018).
Percival, J. M. nNOS regulation of skeletal muscle fatigue and exercise performance. Biophys. Rev. 3, 209–217 (2011).
Pambianco, S. et al. Reversal of defective mitochondrial biogenesis in limb-girdle muscular dystrophy 2D by independent modulation of histone and PGC-1α acetylation. Cell Rep. 17, 3010–3023 (2016).
Cao, Y. et al. S-nitrosoglutathione reductase-dependent PPARγ denitrosylation participates in MSC-derived adipogenesis and osteogenesis. J. Clin. Invest. 125, 1679–1691 (2015).
Cao, Y., Balkan, W. & Hare, J. M. S-nitrosylation and MSC-mediated body composition. Oncotarget 6, 28517–28518 (2015).
Dawlaty, M. M. et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175 (2011).
Liu, L. et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 (2001).
Forrester, M. T., Foster, M. W., Benhar, M. & Stamler, J. S. Detection of protein S-nitrosylation with the biotin switch technique. Free Radic. Biol. Med. 46, 119–126 (2009).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We are very grateful to Vanda Turcanova for lab assistance and Laila Fisher for secretarial work. This work has been in part supported by grants from the Danish Cancer Society (KBVU R72-A4647 and R146-A9414 to G.F.; R146-A9364 to F.C.); AIRC (IG2017 n. 20719 to G.F.; IG2017 to F.C.); NovoNordisk (7559 and 22544 to F.C.); Lundbeckfonden (R233-2016-3360 to F.C.); LEO Foundation (LF17024 to F.C.); Fondazione Roma (to A.M.); AFM-Telethon (to M.T.C.). We also thank the “Uncovering excellence” Grant 2014 [MDESMPLAT to G.F.] funded by Tor Vergata University. Cell Stress and Survival Unit is part of the Center of Excellence for Autophagy, Recycling and Disease (CARD), funded by the Danmarks Grundforskningsfond (DNRF125).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Melino
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Montagna, C., Rizza, S., Cirotti, C. et al. nNOS/GSNOR interaction contributes to skeletal muscle differentiation and homeostasis. Cell Death Dis 10, 354 (2019). https://doi.org/10.1038/s41419-019-1584-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-019-1584-3
This article is cited by
-
Autophagy guards tendon homeostasis
Cell Death & Disease (2022)