Abstract
HSP60 is a mitochondrial localized quality control protein responsible for maintaining mitochondrial function. Although HSP60 is considered both a tumor suppressor and promoter in different types of cancer, the role of HSP60 in human pancreatic ductal adenocarcinoma (PDAC) remains unknown. In this study, we demonstrated that HSP60 was aberrantly expressed in human pancreatic cancer tissues and cell lines. Analysis of the Cancer Genome Atlas database revealed that HSP60 expression is positively correlated with pancreatic cancer. Further, knockdown of HSP60 attenuated pancreatic ductal cancer cell proliferation and migration/invasion, whereas ectopic expression of HSP60 increased tumorigenesis. Using an in vivo tumorigenicity assay, we confirmed that HSP60 promoted the growth of pancreatic ductal cancer cells. Functional analyses demonstrated that HSP60 plays a key role in the regulation of mitochondrial function. Mechanistically, both HSP60 knockdown and oxidative phosphorylation (OXPHOS) inhibition by metformin decreased Erk1/2 phosphorylation and induced apoptosis and cell cycle arrest, whereas Erk1/2 reactivation with EGF promoted cell proliferation. Intriguingly, in vitro ATP supplementation partially restored Erk1/2 phosphorylation and promoted proliferation in PDAC cells with HSP60 knockdown and OXPHOS inhibition. These results suggest that mitochondrial ATP is an important sensor of Erk1/2 regulated apoptosis and the cell cycle in PDAC cells. Thus, our findings indicate for the first time that HSP60 may serve as a novel diagnostic target of human pancreatic cancer, and that inhibition of mitochondrial function using drugs such as metformin may be a beneficial therapeutic strategy targeting pancreatic cancer cells with aberrant function of the HSP60/OXPHOS/Erk1/2 phosphorylation axis.
Similar content being viewed by others
Introduction
Mitochondrial functions, particularly oxidative phosphorylation (OXPHOS), are monitored by several hierarchical quality control (QC) machineries1. Disturbing of mitochondrial QC proteins have been associated with a number of diseases2,3. HSP60 is a mitochondrial matrix localized QC proteins in eukaryote cells. Changes of HSP60 function results in mitochondrial dysfunction and is closely associated with cancer4. Inhibition of HSP60 activity with myrtucommulone induces mitochondrial-mediated cancer cell apoptosis. Because HSP60 is a dual regulator of apoptosis, it has been considered both a tumor suppressor and promoter in different cancer types5,6.
Pancreatic ductal adenocarcinoma (PDAC) is one of the leading causes of death among all cancers worldwide7. Because of its late diagnosis and very poor prognosis, the mortality of pancreatic cancer is almost equal to its incidence. In China, the incidence of pancreatic cancer continually increased from 2000 to 20118. Recently, multiple metabolic reprogramming profiles including the Warburg phenotype, the reverse Warburg phenotype, the glutaminolysis phenotype, and the lipid-dependent phenotype were stratified into different subsets of PDAC cells9. Although mitochondria play a central role in the regulation of metabolic flux, aberrant regulation of mitochondrial functions has been associated with PDAC10. Sustained OXPHOS function with high-mobility group box 1 (HMGB1) 11, the MYC proto-oncogene/ PPARgamma coactivator 1 alpha (PGC-1α) axis12, and receptor for advanced glycation endproducts (RAGE) (also known as AGER) have been associated with poor prognosis of PDAC11. Despite imbalanced adenosine triphosphate (ATP) generation being central to cancer cell fate decision, the underlying mechanism is not fully understood11.
Proteomics analysis has identified several potential protein biomarkers; however, whether there is altered expression of HSP60 in PDAC and normal tissues is not clear. To explore the mechanisms of QC proteins in PDAC, we performed a bioinformatics analysis of QC transcriptomes and discovered that HSP60 sustained mitochondrial function driving the development of PDAC. We found that HSP60 regulated the generation of mitochondrial ATP, which is critical for Erk1/2 (a ras-dependent extracellular signal-regulated kinase) derived anti-apoptotic and cell survival in PDAC cells. In addition, we demonstrated that the mitochondrial respiratory inhibitor metformin decreased Erk1/2 phosphorylation and induced apoptosis and cell cycle arrest in PDAC cells partially through decreased mitochondrial ATP generation. Our current study uncovered a mechanism in which HSP60 promotes cancer cell growth revealing a potential therapeutic strategy targeting mitochondrial respiration in PDAC.
Results
Mitochondrial QC protein Hsp60 modulates tumorigenicity in PDAC
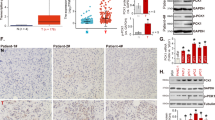
To investigate correlations between mitochondrial QC machinery and PDAC, we performed bioinformatics analysis in PDAC using the Cancer Genome Atlas (TCGA) database. Of the 19 most studied mitochondrial QC proteins (MQCPs), HSP60 (also known as HSPD1) was the only MQCP that had not only significantly increased expression in PDAC tissues (1.58-fold greater) compared with that of normal tissue, but was also positively correlated with PDAC histological grade (correlation coefficient = 0.91, P = 0.006) (Figs. 1a, b and Table 1). Consistently, protein analysis in both paraffinized PDAC and fresh tissue samples confirmed that HSP60 was elevated in cancerous tissues (Figs. 1c, d). Additionally, we found increased HSP60 in PDAC cell lines both with and without KRAS (a Kirsten ras oncogene homolog from the mammalian ras gene family) mutation (Figs. 1e, f, respectively) compared with that found in normal pancreatic ductal cells (hTERT-HPNE). As a related chaperone protein with HSP60, HSP10 (also known as HSPE1) was significantly increased in PDAC tissues compared with normal tissues with the same fold change observed with HSP60; however, expression of HSP10 in PDAC tissues was not correlated with histological grade (Table 1). These findings indicate that HSP60 expression is related to PDAC and that the relationship is independent of KRAS status.
a Analysis of relative HSPD1 expression in four normal pancreas tissues and 176 PDAC tissues from the TCGA database. b Normal and tumor samples were grouped by histological grade: G0 (n = 4), G1 (n = 31), G2 (n = 96), and G3 (n = 49). The column shows a dot map of HSPD1 expression in pancreatic cancer with different histological grades. CC correlation coefficient. c Representative photomicrographs of HSP60 immunohistochemistry (100× and 400×) of normal pancreas and cancerous tissues as well as quantitative immunohistochemistry results of its expression. d A representative western blot showing HSP60 protein expression in pancreas normal (N) and matched PDAC tissue (T). e, f Western blotting analysis of HSP60 protein expression in PDAC cell lines including cell lines e with and f without KRAS mutation. g Cell proliferation and h clone formation assays showing decreased proliferation in HSP60 shRNA-transfected (shHSP60) Panc-1 cell lines compared with that found in control cells. i The migration (upper panel) and invasion (low panel) abilities of control and shHSP6060 Panc-1 cells were evaluated using a transwell assay. j The migration ability of control cells, shHSP60 Panc-1 cells, and rescued (ectopic) expression of HSP60 cells was evaluated using a wound-healing assay. k Tumor xenograft experiments were carried out using shHSP60 Panc-1 cells compared with control cells and cells with rescued HSP60 expression. Tumor volume and weight were calculated for each group (n = 6) at the indicated times after cancer cell injection, and statistical comparisons between the differences were assessed. All quantified data are presented as mean ± SEM (n ≥ 3). *P < 0.05, **P < 0.01, and ***P < 0.001
To characterize the impact of HSP60 on PDAC, we established two Panc-1 cell line models with stable knockdown (KD) of HSP60 at the level of 50% and 80%, respectively (Supplementary Figure S1A and B). Further, ectopic expression of HSP60 in cells from one of the HSP60 KD cell lines restored HSP60 expression (Supplementary Figure S1C). We found that HSP60 KD diminished in vitro PDAC cell proliferation and clonal formation, whereas rescued expression of HSP60 increased the viability of cancer cells (Fig. 1g). Additionally, HSP60 KD significantly reduced invasion (Fig. 1i, lower panel) and migration of cancer cells (Fig. 1i, upper panel, and Fig. 1j), whereas ectopic expression of HSP60 in HSP60 KD cancer cells reversed cancer cell migration (Fig. 1j). Furthermore, using an in vivo tumorigenicity assay, we found that both tumor weight (Fig. 1k, upper panel) and volume (Fig. 1k, lower panel) in mice with HSP60 KD cells were significantly lower compared with control Panc-1 cells, whereas rescued expression of HSP60 in HSP60 KD cells increased tumor formation (Fig. 1k). Taken together, our findings indicate that HSP60 expression modulates tumorigenicity, including cancer cell proliferation, invasion, and migration.
HSP60 maintains mitochondrial function in PDAC cells
Although remodeling of OXPHOS and glycolysis has been frequently observed in multiple cancer types, we analyzed the expression of genes involved in OXPHOS (83 genes), the TCA cycle (16 genes), and glycolysis (10 genes) in PDAC samples from the TCGA database (Supplementary Figure S2A), and we found that the expression levels of two glycolytic genes (HK2 and PKM2), one TCA cycle gene (IDH1), and two OXPHOS-related genes (COX6B2 and NDUFAB1) were positively correlated with histological grade score (Supplementary Figure S2B-F). Further, RNA levels of COX6B2 and NDUFAB1 were also significantly upregulated in PDAC tissues compared with normal pancreatic ductal tissues from the TCGA database (Supplementary Figure S2G and H). Although PDAC cells may have different metabolic profiles9, it is likely that OXPHOS function is increased, or at the least is not decreased, in PDAC cells compared with normal pancreatic cells.
Considering the important role of HSP60 in the regulation of mitochondrial function and that OXPHOS may play a role in PDAC, we tested the effect of HSP60 on OXPHOS in PDAC cells. First, we evaluated mitochondrial function by measuring mitochondrial respiration and ATP content in HSP60 KD cells, and found that all basal mitochondrial respiration, uncoupled mitochondrial respiration, and coupled mitochondrial respiration were significantly decreased compared with control cells (Fig. 2a). In contrast, expression of ectopic HSP60 in HSP60 KD cells partially restored mitochondrial respiration (Fig. 2b). Furthermore, KD of HSP60 decreased mitochondrial ATP content, whereas rescued expression of HSP60 increased mitochondrial ATP content (Fig. 2c).
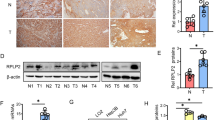
a, b Analysis of the oxygen consumption rate (OCR) of various cell lines by an Oxygraph-2k comparing shHSP60 Panc-1 cells with a control cells and b rescued expression of HSP60 cells. OCR was first measured of each cell line under basal conditions followed by sequential addition of oligomycin (100 μg/mL) at the indicated ATP-linked OCR. Basal, basal OCR; Oligomycin, OCR in the presence of 100 μg/mL oligomycin; Basal-oligomycin, OXPHOS-associated OCR was calculated by subtracting the OCR in the presence of oligomycin from basal OCR. c Measurements of mitochondrial ATP levels. Cells were incubated with 10 mM glucose or 5 mM 2-DG plus 5 mM pyruvate to determine mitochondrial ATP generation. Relative average ATP levels in mitochondria per cell line are shown. d Respiratory chain supercomplexes and single complexes in shHSP60 Panc-1 cells and control cells. e The respiratory chain supercomplex in shHSP60 Panc-1 cells and cells with rescued expression of HSP60. f Mitochondrial ATP levels and g respiratory chain supercomplexes were measured in cells from PDAC cell lines (SW1990, Patu-8988, and Panc-1) compared with normal pancreatic ductal cells (hTERT-HPNE). h Respiratory chain supercomplexes in fresh PDAC tissue samples compared with those found in paired normal tissues. The values of OCR and ATP generation were normalized to protein concentration. Blots were probed with anti-Grim19, anti-UQCRC2, anti-SDHA or anti-COX1, and anti-cyt c for complex I (CI), complex II (CII), complex III (CIII), and complex IV (CIV), respectively. All data are presented as mean ± SEM (n ≥ 3). *P < 0.05, **P < 0.01, and ***P < 0.001
HSP60 is a chaperone protein indispensable for mitochondrial respiratory chain complex assembly. Thus, we next asked if HSP60 KD downregulated the steady-state level of the mitochondrial respirasome in PDAC cells. As shown in the left panel of Fig. 2d, mitochondrial supercomplexes decreased by 40% and 70% in Panc-1 cells with of 50% and 80% of HSP60 KD, respectively. Consistently, cells with HSP60 KD exhibited decreased single mitochondrial respiratory chain complex I, III, and IV (Fig. 2d, right panel). In contrast, rescued expression of HSP60 restored the steady-state level of mitochondrial supercomplexes (Fig. 2e). Similarly, PDAC cells of SW1990, Patu-8988, and Panc-1, all of which express higher levels of HSP60 compared with normal pancreatic ductal cells, generated more mitochondrial ATP and exhibited a greater number of mitochondrial supercomplexes compared with that found in cells from the normal pancreatic ductal cell hTERT-HPNE (Figs. 2f, g). Our findings related to increased HSP60 and respiratory chain supercomplexes were independently verified in three fresh PDAC tissue samples compared with paired normal tissues (Figs. 1d,2h). Taken together, these results suggest that HSP60 regulates mitochondrial function in PDAC cells.
HSP60 promotes the phosphorylation of Erk1/2 via upregulation of OXPHOS
As altered OXPHOS function has been linked to multiple mitochondrial-to-nucleus retrograde signaling pathways, we next investigated several key pathways involved in mitochondrial-to-nucleus signaling in cells from two HSP60 KD Panc-1 cell lines and one HSP60 KD cell line with rescued ectopic expression of HSP60. Of the six major mitochondrial-related pathways, decreased Erk1/2 phosphorylation was observed in HSP60 KD cells compared with control cells (Fig. 3a), whereas Erk1/2 phosphorylation was increased in KD cells with rescued ectopic expression of HSP60 (Fig. 3b). Consistently, we found that the increased HSP60 observed in KRAS wild-type and mutant PDAC cells compared with hTERT-HPNE cells (Fig. 1f) was also associated with increased phosphorylation of Erk1/2 compared with that of normal pancreatic ductal cells (Fig. 3c and Supplementary Figure S3). Similarly, fresh PDAC tissues, which exhibited increased levels of HSP60, also showed higher levels of phosphorylated Erk1/2 (Figs. 1d, 3d). These results show that HSP60 overexpression in PDAC cells may be associated with Erk1/2 phosphorylation.
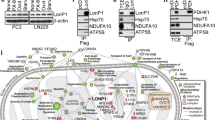
a Analysis of mitochondrial-to-nucleus pathways in shHSP60 Panc-1 cells and control (Ctrl) cells. Cell lysates were analyzed by western blot with antibodies against P38, p-p38, Src, p-Src, Akt1, p-Akt (Thr308), p-Akt (Ser473), Erk1/2, p-Erk1/2, AMPKα, p- AMPKα, JNK, p-JNK, and actin. b–d Western blot analysis of p-Erk1/2 in the indicated cells and tissues: b shHSP60 Panc-1 cells vs. cells with rescued expression of HSP60, c cells from PDAC cell lines (SW1990, Patu-8988, and Panc-1) vs. adjacent normal pancreatic ductal cells (hTERT-HPNE), and d fresh PDAC tissue vs. paired normal tissue. N adjacent normal tissue; T tumor tissue
We next asked whether decreased Erk1/2 phosphorylation in HSP60 KD cells was mediated by the attenuated OXPHOS function. As shown in Figs. 4a, b, inhibition of OXPHOS by either rotenone or Ethidium bromide (EB, inhibitor of mitochondrial DNA replication) reduced Erk1/2 phosphorylation in Panc-1 cells. To determine whether mitochondrial membrane potential (MMP) was involved in Erk1/2 phosphorylation, cells were treated with carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) and oligomycin to depolarize and hyperpolarize the MMP, respectively. Unexpectedly, we found that both FCCP and oligomycin treatment decreased Erk1/2 phosphorylation (Fig. 4c). Additionally, although HSP60 KD caused oxidative stress in mitochondria (Supplementary Figure S4A), treatment using NAC in Panc-1 cells did not increase Erk1/2 phosphorylation in HSP60 KD cells (Supplementary Figure S4B). Because HSP60 KD impairs assembly of the respirasome followed by decreased mitochondrial ATP generation, we next determined whether the ATP generated from mitochondria is important for Erk1/2 phosphorylation. Antagonism of ATP translocation from mitochondria into the cytosol using bongkrekic acid (BKA), an inhibitor of the adenine nucleotide translocator (ANT), revealed that decreased ATP content in the cytosol inhibited Erk1/2 phosphorylation, a finding that indicates the ATP generated from OXPHOS is indispensable for Erk1/2 activation (Fig. 4d). Similarly, ATP complementation in HSP60 KD cells partially restored Erk1/2 phosphorylation in a time-dependent manner (Fig. 4e).
a–c Inhibition of OXPHOS significantly affects the phosphorylation of Erk1/2. Western blot analysis of the effects of: a rotenone on the phosphorylation of Erk1/2 in cells treated with rotenone (200 nM) for 24 h, b Ethidium bromide (EB, an inhibitor of mitochondrial DNA replication) on the phosphorylation of Erk1/2 in cells treated with 50 μg/mL EB for 6–8 days, and c oligomycin on the phosphorylation of Erk1/2 in cells treated with 80 μM oligomycin for 24 h. d, e Mitochondrial ATP content significantly affects the phosphorylation of Erk1/2. Western blot analysis of the effects of d, Bongkrekic acid (BKA), an inhibitor of the adenine nucleotide translocator (ANT), on the phosphorylation of Erk1/2 in cells treated with BKA (0, 6, 8, 10, and 12 μM) for 24 h, and e ATP on the phosphorylation of Erk1/2 in shHSP60 Panc-1 cells treated with 10 mM ATP for 1 and 2 min. β-Actin was used as an internal control. f Western blot analysis of cytosol and mitochondrial p-Erk1/2 levels in Panc-1 control (Ctrl) cells treated with 80 µM oligomycin and shHSP60 Panc-1 cells compared with Ctrl cells. β-Actin and VDAC were used as cytosol and mitochondrial controls, respectively. g Colocalization of p-Erk1/2 to cytosol and mitochondria. Ctrl and shHSP60 Panc-1 cells were probed with p-Erk1/2 (green) and mitochondria (MitoTracker Red). All cells were background stained with DAPI (blue). The images were taken with a confocal laser microscope and representative images (600×) are shown
As Erk1/2 is localized in both the outer mitochondrial membrane and the cytosol, we examined the status of Erk1/2 phosphorylation in both sites and found that inhibition of OXPHOS by oligomycin in Panc-1 cells decreased Erk1/2 phosphorylation in both mitochondria and cytosol (Fig. 4f). Similarly, HSP60 KD also suppressed Erk1/2 phosphorylation in both mitochondria and cytosol (Figs. 4f, g). These results indicate that HSP60 maintains OXPHOS function and promotes mitochondrial ATP generation, which in turn promotes Erk1/2 phosphorylation in PDAC cells.
Metformin attenuates cancer cell growth via decreased mitochondrial ATP generation and subsequent Erk1/2 phosphorylation
Although the underlying mechanism is unclear, PDAC cell growth is inhibited by the anticancer drug candidate metformin. Because it has been demonstrated that metformin inhibits mitochondrial complex I activity13, we speculated that metformin may prevent Erk1/2 phosphorylation via decreased ATP generation to attenuate cancer cell growth. To test this hypothesis, we determined the level of the mitochondrial respirasome and confirmed that metformin impaired mitochondrial supercomplex assembly (Fig. 5a), through which mitochondrial ATP generation was dramatically decreased (Fig. 5b). Consistent with these findings, we also found that inhibition of mitochondrial ATP generation by metformin decreased Erk1/2 phosphorylation both in mitochondria and cytosol (Fig. 5c). In addition, ATP supplementation in cells treated with metformin partially rescued Erk1/2 phosphorylation (Fig. 5d). These results show that metformin inhibits Erk1/2 phosphorylation to some extent through downregulation of mitochondrial ATP generation. Furthermore, inhibiting Erk1/2 phosphorylation in control Panc-1 cells by U0126 (an inhibitor of the mitogen-activated protein (MAP) kinase kinases, MEK1 and MEK2) or metformin (Supplementary Figure S5 and Fig. 5c) mimics the effect of clonal formation found in HSP60 KD cells compared with control Panc-1 cells (Fig. 5e). Moreover, treatment with EGF, which is an inducer of Erk1/2 phosphorylation, promoted clonal formation in both HSP60 KD and metformin-treated Panc-1 cells (Figs. 5f, g), findings that support Erk1/2 phosphorylation has a role in HSP60 KD-related and metformin-induced cancer cell growth inhibition. Of note, cells treated with metformin formed fewer clones than those cells incubated with U0126 (Fig. 5e). Furthermore, reactivated Erk1/2 did not fully restore clonal formation in Panc-1 cells treated with metformin (Fig. 5g). This finding indicates that the tumor inhibitory effects of metformin are partially mediated through regulation of Erk1/2 phosphorylation. Together, these findings indicate that decreased Erk1/2 phosphorylation in HSP60 KD inhibits cancer cell growth and that OXPHOS inhibitors such as metformin may be used in the treatment of PDAC exhibiting increased Erk1/2 phosphorylation.
a Mitochondrial ATP levels and b assembly of respiratory chain supercomplexes were determined in Panc-1 cells treated with 10 mM metformin (Met) for 72 h. c Western blot analysis of cytosol and mitochondrial p-Erk1/2 levels in Panc-1 (Ctrl) cells treated with 10 mM Met for 72 h. β-Actin and VDAC were used as cytosol and mitochondrial controls, respectively. d Western blot analysis of cytosol and mitochondrial p-Erk1/2 levels in Met pre-treated (72 h) Panc-1 (Ctrl) cells with and without ATP at concentrations of 150 nM and 200 nM for 5 min. e Clonal formation assay of Panc-1 Ctrl cells, HSP60 KD cells, and Panc-1 Ctrl cells treated with 10 mM Met and 50 μM U0126 for 24 h. f, g Clonal formation assay of f HSP60 KD and g Met-treated (10 mM) Panc-1 cells with or without EGF (30 ng/mL). U0126 is an inhibitor of the MAP kinase kinases, MEK1 and MEK2. Data are presented as mean ± SEM (n ≥ 3). **P < 0.01 and ***P < 0.001
HSP60 KD and metformin administration induce apoptosis and cell cycle arrest in PDAC cells
To gain further insights into the effects of HSP60 KD and metformin on the inhibition of tumorigenesis, we next performed RNA-sequencing in wild-type Panc-1 cells, HSP60-silenced Panc-1 cells, and metformin-treated Panc-1 cells. We identified 8997 and 7104 differentially expressed genes (DEGs) in HSP60-silenced and metformin-treated PDAC cells, respectively (Fig. 6a). Of the 5285 common DEGs, we identified 4609 of which 1681 were downregulated and 2928 were upregulated in both HSP60-silenced and metformin-treated PDAC cells (Fig. 6a). Pathway enrichment analysis of the 4609 shared DEGs using Gene Ontology (GO) revealed a large proportion of significantly enriched pathways clustered into specific cell cycle-related processes, such as DNA replication, DNA repair, cell cycle check point, and ribosome biogenesis (Fig. 6b). Considering the fact that Erk1/2 plays an essential role in cell fate decisions, downstream effectors of Erk1/2 in mitochondrial apoptosis and the cell cycle were analyzed. As shown in Fig. 6c, we found drastically increased expression of mitochondrial apoptosis genes, including BAX, BAK1, and BID, in both HSP60 KD and metformin-treated PDAC cells, of which protein level of BAX in control, HSP60 KD, and metformin-treated PDAC cells was further confirmed (Fig. 6d). In addition, cell cycle negative regulators, such as CDKN1A, RBL1, ATR, and TP53, were upregulated (Fig. 6c). Consistently, HSP60 KD and metformin treatment induced G1 cell cycle arrest and apoptosis in Panc-1 cells (Figs. 6e, f). These findings suggest that both metformin- and HSP60 KD-induced PDAC cell growth inhibition is mediated by inhibition of Erk1/2 phosphorylation and activity of its downstream effectors.
a DEGs were identified in HSP60 KD and metformin-treated (10 mM for 72 h) Panc-1 cells. b Pathway enrichment analysis of 4,609 shared DEGs using the GO database. c Heat map showing hierarchical clustering of apoptosis and cell cycle genes related to p-Erk1/2 in HSP60 KD and metformin-treated (10 mM for 72 h) Panc-1 cells. d A representative western blot showing HSP60 protein level in control Panc-1 cells, HSP60 KD, and metformin-treated (10 mM for 72 h) Panc-1 cells. e Cell cycle stage and f apoptosis were determined by flow cytometry analysis in control Panc-1 cells, HSP60 KD, and metformin-treated (10 mM for 72 h) Panc-1 cells. The column shows the percentages of cells at d different phases and e the percentage of apoptotic (early and late) cells. Data are presented as mean ± SEM (n ≥ 3). *P < 0.05 and ***P < 0.001
Discussion
To cope with the high protein turnover rate, QC proteins that monitor the folding of newly synthesized proteins and their subsequent assembly, as well as the disassembly of damaged proteins for degradation are over activated in multiple rapidly growing cancer cells14. However, activated chaperones such as HSP90 are also indispensable to suppress cancer cell growth by stabilizing tumor suppressors, such as P53 and VHL15. In mitochondria, QC proteins including heat shock family proteins and ATP-dependent proteases not only maintain mitochondrial proteostasis to protect the cells from stress induced apoptosis16, but also reprogram metabolism to facilitate cancer cell growth17. Of these QC proteins, HSP60 is one the most studied chaperones in cancer biology18. Similar to HSP90, HSP60 functions as either a tumor suppressor or promoter in different cancer types5,19. On the one hand, proteomics analysis identified HSP60 as a cancer-associated protein marker in both colorectal and breast cancers20,21, and in addition to mitochondrion, increased expression of HSP60 was found in different subcellular structures22. On the other hand, HSP60 may serve as a tumor suppressor in hepatocellular and clear cell renal cell carcinoma5,23. In PDAC, evaluation of HSP60 expression in tumor was confounded by different detection methods24,25. In the current study, we demonstrated that HSP60 expression was significantly increased in cancer tissues compared with that of adjacent normal tissues. Although we found no evidence of correlation between the histological grade and HSP60 expression in our cohort of patients with PDAC, we found that HSP60 expression was positively correlated with disease severity from the TCGA database. The reason for this discordant finding may be because of the limited local sample size. Considering that HSP60 and HSP10 are co-chaperones working together in mitochondria26, a coordinated expression pattern of these two proteins were frequently observed in different cancer types27,28 including PDAC (Table 1). Although more data are required to clarify the effects of HSP60 expression on cancer progression, our study provides the first description of increased HSP60 expression in PDAC.
As a key component of mitochondrial QC machinery, HSP60 guards mitochondrial function under multiple nutrition stress3. Constitutive expression of HSP60 in mitochondria is involved in stabilizing mitochondrial DNA(mtDNA) nucleoids and newly synthesized mtDNA encoded polypeptides29,30, indicating a pivotal role of HSP60 in the maintaining of OXPHOS function. In this study, we confirmed that HSP60 repression inhibited mitochondrial electron transfer and subsequent ATP synthesis, whereas elevated HSP60 boost the mitochondrial function. Generally, QC proteins including HSP60 ensure the proper mitochondrial protein folding to support the mitochondrial function at the protein level31, both loss/decreased of QC function and accumulation of misfolded mitochondrial protein that beyond the regulatory capacity of mitochondrial unfolded protein response, can activate organelle QC via mitophagy or mitochondrial fission and fusion32. Notably, HSP60 was shown to interact with RNA-binding protein FUS/TLS, and facilitate the mitochondrial fragmentation in addition to its QC property33. Although we did not look into the detail of mitochondrial fragmentation in this study, changes of mitochondrial mass was likely not affected in HSP60 KD cells (Fig. 4g). Additionally, HSP60 KD may change the mitochondrial cristae shape, which is important for mitochondrial supercomplex assembly34. Furthermore, HSP60 KD may also disturb the mitochondrial supercomplex assembly by impairment of newly synthesized OXPHOS subunits folding through HSP10/HSP60 complex35. However, further studies are required to clarify how HSP60 KD induced mitochondrial supercomplex assembly.
Although disturbances of mitochondrial function alter mitochondrial retrograde signaling36, altered expression of mitochondrial QC proteins was shown to boost cancer-related signaling pathways because of their pivotal roles in maintaining OXPHOS machinery37. HSP60 activates many inflammatory related signaling pathways, but few of them are mitochondrial-related38. Recently, HSP60 KD in renal cancer was shown to induce Reactive oxygen species (ROS) accumulation and subsequent activation of the adenosine monophosphate-activated protein kinase (AMPK) pathway. In our study, we found ROS accumulation in PDAC cells with HSP60 KD, but AMPK was not significantly affected (Fig. 3a). Instead, we found that HSP60 was positively related with Erk1/2 phosphorylation via HSP60-induced cytokine secretion39. Our study reports for the first time that HSP60 is important for the activation of mitochondrial retrograde Erk1/2 phosphorylation. Previous studies have shown that HSP60 functions as a tumor promoter by directly interacting with apoptosis/anti-apoptosis-related proteins to determine cancer cell fate. For example, HSP60 directly interacts with and inhibits xc of Bcl-xl, which is an anti-apoptotic mitochondrial outer membrane protein40. Further, it interacts with and stabilizes survivin to attenuate caspase-dependent apoptosis41, and forms a complex with clusterin to promote cancer cell survival and proliferation42. In this study, we uncovered a second mechanistic pathway of HSP60-regulated anti-apoptosis via an HSP60/OXPHOS/Erk1/2 axis.
Functional OXPHOS is indispensable to support cancer cell growth via selective activation of cancer-related mitochondrial retrograde signaling rather than energy supplementation43. Mitochondria contain multiple signal mediators, of which ATP is a sensor molecular for many pathways. Although Warburg effect is frequently observed in many cancer types, sustained OXPHOS function and subsequent ATP generation is vital in PDAC cells11,44. Our results showed that HSP60 KD in PDAC cells reduce mitochondrial ATP generation and induces cell cycle arrest and apoptosis, whereas rescued ectopic expression of HSP60 in HSP60 KD cells re-boosts mitochondrial ATP generation and promotes cell proliferation. The effects of intracellular ATP generated from glycolysis on apoptosis were previously demonstrated45. In vitro supplementation of ATP was shown to promote Erk1/2 phosphorylation46. However, we show here that HSP60 KD lowers endogenous mitochondrial ATP generation and activates mitochondrial apoptosis by reducing Erk1/2 phosphorylation in PDAC cells. Similarly, a causal relationship between mitochondrial ATP and ERK1/2 activation was also claimed in an ischemia/reperfusion model47. Furthermore, we confirmed that in vitro ATP supplementation promoted Erk1/2 phosphorylation in HSP60 KD cells, whereas downregulation of OXPHOS function in PDAC cells with metformin decreased Erk1/2 phosphorylation and increased Erk1/2-related apoptosis. The effect of metformin on the inhibition of Erk1/2 phosphorylation has been proposed48; however, the underlying mechanism of metformin in the modulation of ERK1/2 is poorly understood. Under this context, we described in the present study an unconventional role of metformin in PDAC in which the administration of metformin caused overexpression of HSP60, increased activation of OXPHOS, and elevated Erk1/2 phosphorylation in PDAC cells. Although further studies are crucial to determine whether upstream effectors of Erk1/2 or Erk1/2 itself are regulated by mitochondrial ATP, OXPHOS inhibitors such as metformin may serve as potential drugs for the treatment of PDAC. In addition, further investigations are warranted to test the effectiveness of molecules such as metformin in PDAC cells with different levels of HSP60 expression and OXPHOS activity.
In summary, our findings demonstrated for the first time that HSP60 promotes cancer cell proliferation and serves as an anti-apoptotic mediator in PDAC. Investigation in the underlying mechanisms revealed that HSP60 maintains OXPHOS function to generate ATP, which is essential for mitochondrial and cytosol Erk1/2 phosphorylation. Meaningfully, this improved understanding of the mechanism supports the use of anticancer agents such as metformin to target mitochondrial ATP generation in PDAC cells with high levels of HSP60 expression, OXPHOS activity, and Erk1/2 phosphorylation.
Materials and methods
Cell lines and culture conditions
Pancreatic cancer cells SW1990 and BXPC-3 were ordered from Cell Resource Center, Chinese Academy of Medical Sciences in 2017. Pancreatic cancer cells Panc-1 and Patu-8988 were ordered from the same center in 2015. Test of the cells authentication were exempted for these cells from Cell Resource Center, Chinese Academy of Medical Sciences. Immortalized pancreatic ductal epithelium cells (hTERT-HPNE) was gifted by BaiRong Biotechnology (Shanghai, China) in 2017, and authenticated using short tandem repeat (STR) profiling analysis by the same center in August of 2017. All cells were cultured in high-glucose Dulbecco’s modified Eagle's medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% cosmic calf serum (Sigma-Aldrich, St. Louis, MO, USA).
Patients and PDAC tissue specimens
Twenty-eight pairs of 5-μm-thick formalin-fixed paraffin-embedded sections of primary PDAC specimens and paired adjacent normal pancreas tissues were obtained from patients at the First Affiliated Hospital of Wenzhou Medical University (Zhejiang, China). Three pairs of matched PDAC tumor tissues and adjacent normal tissues from each patient were immediately snap frozen in liquid nitrogen after surgical removal at the First Affiliated Hospital of Wenzhou Medical University. Informed consent was obtained from all subjects under protocols approved by the Ethical Committee of the First Affiliated Hospital of Wenzhou Medical University. All experimental methods were carried out in accordance with approved guidelines of Wenzhou Medical University.
Immunohistochemistry
Tissue sections were mounted on slides, treated with xylene, and rehydrated using a graded series of alcohol–water solutions before heat treatment in 10 mM citric acid (pH 6.0) for antigen retrieval49. Endogenous peroxidase activity was quenched using a 0.3% H2O2 solution. After blocking with 10% serum, sections were incubated at 4 °C overnight with primary anti-HSP60 antibodies (sc-376261; 1:200; Santa Cruz Biotechnology, Dallas, TX, USA). The slides were then incubated with anti-mouse IgG antibody (ZSBIO, Beijing, China) at 37 °C for 30 min. Signals were visualized with diaminobenzidine (DAB) (ZSBIO), counterstained with hematoxylin, dehydrated in ethanol, cleared in xylene, and mounted. Protein expression levels were analyzed by calculating the integrated optical density per stained area (IOD/area, mean OD value, MOD) using Image-Pro Plus 6.0 (Media Cybernetics, Rockville, MD, USA) as described previously50.
Antibodies and immunoblotting
Proteins from whole-cell and clinical tissues were extracted with RIPA lysis buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with a protease inhibitor cocktail (Sigma-Aldrich). Cytoplasmic and mitochondrial fractions were isolated from cultured cells as described previously51. Mitochondrial membrane proteins were extracted using digitonin or Triton X-100 (Sigma-Aldrich) as described in our previous study52. Proteins separated by blue native polyacrylamide gel electrophoresis (BN-PAGE) or sodium dodecyl sulfate polyacrylamide gel electrophoresis(SDS-PAGE) were transferred to 0.22 µm polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA) using a semi-dry transfer system (Bio-Rad). Proteins were probed with anti-Hsp60 (sc-376261; 1:1000; Santa Cruz Biotechnology), anti-ERK1/2 (#9102; 1:1000; Cell Signaling Technology), anti-phospho-ERK (Thr202/Tyr204) (#9101; 1:1000; Cell Signaling Technology), anti-AMPKα (#2532; 1:1000; Cell Signaling Technology), anti-phospho-AMPKα (Thr172) (#2535; 1:1000; Cell Signaling Technology), anti-P38 (#9212; 1:1000; Cell Signaling Technology), anti-phospho-P38 (Thr389) (#9211; 1:1000; Cell Signaling Technology), anti-Src (#2109; 1:1000; Cell Signaling Technology), anti-phospho-Src (#2105; 1:1000; Cell Signaling Technology), anti-AKT1 (#2938; 1:1000; Cell Signaling Technology), anti-phospho-AKT (Ser473) (#12694; 1:1000; Cell Signaling Technology), anti-phospho-AKT (Thr308) (#5106; 1:1000; Cell Signaling Technology), anti-JNK (#9252; 1:1000; Cell Signaling Technology), anti-phospho-JNK (#4668; 1:1000; Cell Signaling Technology), anti-NDUFA13 (ab110240; 1:1000; Abcam, Cambridge, MA, USA), anti-SDHA (ab14715; 1:1000; Abcam), anti-core2 (MS304; 1:1000; Abcam), anti-COXI (MS404; 1:1000; Abcam), anti-ATP synthase subunit alpha (ab14748; 1:1000; Abcam), anti-β-actin (sc-47778; 1:5000; Santa Cruz Biotechnology) or anti-VDAC (#4661; 1:1000; Cell Signaling Technology) antibodies, and then incubated with a horseradish peroxidase-conjugated anti-rabbit/mouse IgG (#7074 / #7076; 1:2000; Cell Signaling Technology) secondary antibody. Signals were detected with Super Signal West Pico chemiluminescent substrate (Thermo Fisher Scientific). Integrated optical density quantification was performed using a Gel-Pro Analyzer 4.0 (Media Cybernetics).
Plasmids
HSP60 and control luciferase short hairpin RNA (shRNA) were cloned into the lentivirus plasmid pLKO.1. The shRNA sequences for HSP60 were as follows: shHSP60-1, 5′-TTCAAGAGCAGGTACAATG-3′ (ORF region); shHSP60-2, 5′-CCATCAGTTACTGGTTTCAGT-3′ (3′-UTR region). The shRNA sequence of luciferase was previously described53. For HSP60 overexpression, a full-length HSP60 complementary DNA (cDNA) was synthesized and cloned into pcDNA3.1. Stable cell lines were selected using 800 μg/mL G418 or 4 μg/mL puromycin (Sigma-Aldrich).
Chemicals and reagents
Rotenone, oligomycin, FCCP, EB, ATP, N-acetyl cysteine (NAC), 2-deoxy-d-glucose (2-DG), and pyruvate were purchased from Sigma-Aldrich. BKA and metformin were obtained from Santa Cruz Biotechnology (Dallas, TX, USA) and U0126 was obtained from SelleckChem (Houston, TX, USA).
Cell proliferation assay
A cell proliferation assay was performed using Cell Counting Kit-8® solution (Dojindo, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Briefly, cells were seeded in wells at a concentration of 4 × 103 cells/100 μL in 96-well plates and treated with 10 μL of Cell Counting Kit-8® solution during the last 4 h of culture. Optical density of the wells was measured at 450 nm using a Varioskan™ Flash Multimode Reader (Thermo Fisher Scientific).
Colony formation assay
For colony formation assays, cells were seeded in six-well plates at a density of 1 × 103 cells per well. After 13–15 days, colonies were fixed with 4% paraformaldehyde and stained with crystal violet. Cell colonies whose diameters exceeded 0.5 mm were counted using Gel-Pro Analyzer 4.0 (Media Cybernetics).
Cell migration and motility analysis
Cells were cultured in six-well plates incubated overnight yielding confluent monolayers for the wound-healing test. Wounds were made using a pipette tip and photographs were taken immediately (time zero) and once daily until wound healing. The distance migrated by the cell monolayer to close the wounded area during this time period was measured. The migration of cells into the wound area was quantified by Image J v 2.4.1.7 (National Institutes of Health, Bethesda, MD, USA).
The cell motility assay was performed using a 24-well transwell plate chamber with a pore size of 8 μm (Costar, Cambridge, MA, USA). Cells (2 × 104) were seeded in serum-free media and translocated toward complete growth media. After 24 h of incubation at 37 °C, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of invaded cells was counted in three randomly selected high-power fields under a microscope.
Matrigel invasion assay
The cell invasion assay was performed using a Transwell plate chamber. The inserts were coated with 20 μL Matrigel (1:9 dilution; BD Biosciences, San Jose, CA, USA). Cells (4 × 104) were seeded in serum-free medium and translocated toward complete growth media. After 24 h of incubation at 37 °C, the remainder of the process was identical to that of the motility assay54.
ATP measurement
ATP was measured using an ATP measurement kit (Molecular Probes, Carlsbad, CA, USA) according to the manufacturer’s instructions. To measure mitochondrial ATP, cells were incubated with 10 mM glucose (Sigma-Aldrich) or 5 mM 2-DG with 5 mM pyruvate for 2 h prior to measurement. Fluorescence/luminescence was measured using a Varioskan™ Flash Multimode Reader (Thermo Fisher Scientific).
Measurements of oxygen consumption
Endogenous oxygen consumption from intact cells was determined using a Clark-type oxygen electrode (Oroboros Instruments, Innsbruck, Austria) as described previously55. After recording basal respiration, oligomycin (100 μg/mL) (Sigma-Aldrich) was added to measure un-coupling respiration.
ROS measurement
Approximately 1 × 106 cells were incubated with 6-chloromethyl-2',7'-dichlorodihydrofluorescein diacetate, acetyl ester (carboxy-H2DCFDA) (20 µm) (Thermo Fisher Scientific) for 30 min at 37 °C. Cells were washed and harvested in Hank’s buffered salt solution and fluorescence was record by a Varioskan™ Flash Multimode Reader (Thermo Fisher Scientific) with excitation at 488 nm and emission at 530 nm. Results are presented as the mean of fluorescence intensity.
Immunofluorescence staining
Cells were cultured on coverslips and incubated in DMEM containing 100 nM MitoTracker Red (Invitrogen, Carlsbad, CA, USA) for 30 min at 37 °C in the dark. After staining, cells were washed with fresh growth medium, and fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS) for 20 min. Cells were then permeabilized with PBS containing 0.2% Triton X-100 for 3 min at room temperature, and incubated overnight with anti-HSP60 (1:300; Santa Cruz Biotechnology) or anti-phospho-ERK (Thr202/Tyr204) (1:250; Cell Signaling Technology) antibodies at 4 °C in a darkroom. Next, cells were washed with PBS and incubated with a fluorescently labeled secondary antibody either IgG-Alexa Fluor 594 or IgG-Alexa Fluor 488 (1:300; Cell Signaling Technology) for 1 h at room temperature in the dark. A final incubation was performed to stain the cells with 4,6-diamidino-2-phenylindole (DAPI; Beyotime, Jiangsu, China) for 15 min at room temperature. After mounting the coverslips, images were captured using a confocal laser microscope at a magnification of ×600 (Nikon, Telford, UK).
Apoptosis assay
Apoptosis assays were performed using an Annexin V- Fluorescein isothiocyanate (FITC) apoptosis detection kit according to the manufacturer’s protocol (Keygen Biotech, Jiangsu, China). Briefly, cells (1 × 106) were collected and washed twice with PBS. Annexin V-FITC and propidium iodide (PI) (Keygen Biotech) were added and Fluorescence activated Cell Sorting (FACS) using a BD Accuri™ C6 (BD Biosciences, Ashland, OR, USA) was performed after 15 min of incubation at room temperature.
Cell cycle analysis
Cells (1 × 106) were collected, washed twice with PBS, and fixed with 75% cold ethanol at 4 °C for 12 h. After two washes with PBS, cells were co-incubated with 50 μg/mL propidium iodide (Keygen Biotech) and 10 μg/mL RNaseA (Keygen Biotech) at 4 °C for 30 min in the dark. Cells were then analyzed for DNA content by FACS using a BD Accuri™ C6 (BD Biosciences) and the results of the cell cycle were analyzed by FlowJo (Tree Star, Ashland, OR, USA).
Xenograft experiments
All animal studies were conducted in accordance with the principles and procedures outlined in the institutional animal ethical committee of Wenzhou Medical University. Approximately 4 × 106 Panc-1 cells were injected subcutaneously into 6-week-old female nude mice (Shanghai Laboratory Animal Center, Shanghai, China). Tumor volume (mm3) defined as (length × width2)/2 was measured using a caliper every 3–4 days up to study termination. After 8–9 weeks, all animals were sacrificed to measure the final tumor volume and tumor weight.
Sample preparation and RNA sequencing
Total RNA was isolated from two biological replicates of each group of treated cells using the RNeasy Mini Extraction kit (Qiagen, Valencia, CA, USA). mRNA from 20 μg of total RNA was purified using polyT-attached magnetic beads followed by fragmentation. First- and second-strand cDNA were synthesized in library preparation for transcriptome sequencing. Clustering of index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on a HiSeq 2000 (Illumina).
Sequencing annotation, identification, and pathway enrichment analysis of DEGs
Clean reads were obtained by deleting the adaptor-only sequences and low-quality sequences. Comparison of the sequences was carried out by BLASTN against the 1000 Genomes Build 37 Decoy 5. The number of annotated clean reads of each gene was analyzed and normalized as reads per kilobase per million reads. DEGs were identified as those genes with an adjusted P < 0.05 using DESeq2. GO enrichment analysis of DEGs was implemented by the clusterProfiler R package in which gene length bias was corrected. Pathway enrichment analysis based on GO analysis of DEGs was used to identify significantly enriched metabolic pathways or signal transduction pathways using an adjusted P-value < 0.05 as the threshold for significance.
Statistical analysis
All experiments were performed in triplicate and were performed independently at least three times. Data are presented as the mean ± standard error of the mean (SEM). All statistical analyses were performed with SPSS 21.0 (IBM, Armonk, NY, USA). Significance was estimated using either independent Student’s t-test or one-way analysis of variance (ANOVA). A null hypothesis was rejected when P < 0.05.
References
Rugarli, E. I. & Langer, T. Mitochondrial quality control: a matter of life and death for neurons. EMBO. J. 31, 1336–1349 (2012).
Magen, D. et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am. J. Hum. Genet. 83, 30–42 (2008).
Juwono, J. & Martinus, R. D. Does Hsp60 provide a link between mitochondrial stress and inflammation in diabetes mellitus? J. Diabetes Res. 2016, 8017571 (2016).
Magnoni, R. et al. The Hsp60 folding machinery is crucial for manganese superoxide dismutase folding and function. Free Radic. Res. 48, 168–179 (2014).
Zhang, J. et al. Hsp60 exerts a tumor suppressor function by inducing cell differentiation and inhibiting invasion in hepatocellular carcinoma. Oncotarget 7, 68976–68989 (2016).
Marino Gammazza, A. et al. Doxorubicin anti-tumor mechanisms include Hsp60 post-translational modifications leading to the Hsp60/p53 complex dissociation and instauration of replicative senescence. Cancer Lett. 385, 75–86 (2017).
Vincent, A., Herman, J., Schulick, R., Hruban, R. H. & Goggins, M. Pancreatic cancer. Lancet 378, 607–620 (2011).
Chen, W. et al. Cancer statistics in China, 2015. Ca. Cancer J. Clin. 66, 115–132 (2016).
Daemen, A. et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. USA 112, E4410–E4417 (2015).
Viale, A. et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014).
Kang, R. et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 33, 567–577 (2014).
Sancho, P. et al. MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell. Metab. 22, 590–605 (2015).
Andrzejewski, S., Gravel, S. P., Pollak, M. & St-Pierre, J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2, 12 (2014).
Starenki, D., Hong, S. K., Lloyd, R. V. & Park, J. I. Mortalin (GRP75/HSPA9) upregulation promotes survival and proliferation of medullary thyroid carcinoma cells. Oncogene 34, 4624–4634 (2015).
Walerych, D. et al. Hsp90 chaperones wild-type p53 tumor suppressor protein. J. Biol. Chem. 279, 48836–48845 (2004).
Takayama, S., Reed, J. C. & Homma, S. Heat-shock proteins as regulators of apoptosis. Oncogene 22, 9041–9047 (2003).
Quiros, P. M., Langer, T. & Lopez-Otin, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell. Biol. 16, 345–359 (2015).
Lianos, G. D. et al. The role of heat shock proteins in cancer. Cancer Lett. 360, 114–118 (2015).
Hwang, Y. J. et al. Expression of heat shock protein 60 kDa is upregulated in cervical cancer. Yonsei. Med. J. 50, 399–406 (2009).
He, Y. et al. Proteomics-based identification of HSP60 as a tumor-associated antigen in colorectal cancer. Proteom. Clin. Appl. 1, 336–342 (2007).
Desmetz, C. et al. Proteomics-based identification of HSP60 as a tumor-associated antigen in early stage breast cancer and ductal carcinoma in situ. J. Proteome Res. 7, 3830–3837 (2008).
Campanella, C. et al. The histone deacetylase inhibitor SAHA induces HSP60 nitration and its extracellular release by exosomal vesicles in human lung-derived carcinoma cells. Oncotarget 7, 28849–28867 (2016).
Tang, H. et al. Downregulation of HSP60 disrupts mitochondrial proteostasis to promote tumorigenesis and progression in clear cell renal cell carcinoma. Oncotarget 7, 38822–38834 (2016).
Qi, T. et al. Comparative proteomic analysis for the detection of biomarkers in pancreatic ductal adenocarcinomas. J. Clin. Pathol. 61, 49–58 (2008).
Cui, Y. et al. Proteomic analysis of pancreatic ductal adenocarcinoma compared with normal adjacent pancreatic tissue and pancreatic benign cystadenoma. Pancreatology 9, 89–98 (2009).
Bottinger, L. et al. Mitochondrial heat shock protein (Hsp) 70 and Hsp10 cooperate in the formation of Hsp60 complexes. J. Biol. Chem. 290, 11611–11622 (2015).
Cappello, F. et al. Hsp60 and Hsp10 down-regulation predicts bronchial epithelial carcinogenesis in smokers with chronic obstructive pulmonary disease. Cancer 107, 2417–2424 (2006).
Cappello, F. et al. The expression of HSP60 and HSP10 in large bowel carcinomas with lymph node metastase. BMC Cancer 5, 139 (2005).
Kaufman, B. A., Kolesar, J. E., Perlman, P. S. & Butow, R. A. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J. Cell. Biol. 163, 457–461 (2003).
Yin, W. et al. Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke 39, 3057–3063 (2008).
Baker, M. J., Tatsuta, T., Langer, T. Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 2011, 3.
Senft, D. & Ronai, Z. A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 40, 141–148 (2015).
Deng, J. et al. FUS interacts with HSP60 to promote mitochondrial damage. PLoS. Genet. 11, e1005357 (2015).
Cogliati, S. et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171 (2013).
Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell. Biol. 14, 630–642 (2013).
Wallace, D. C. Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 (2012).
Seo, J. H. et al. The mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol. 14, e1002507 (2016).
Zanin-Zhorov, A. et al. Heat shock protein 60 enhances CD4+CD25+ regulatory T cell function via innate TLR2 signaling. J. Clin. Invest. 116, 2022–2032 (2006).
Marker, T. et al. Heat shock protein 60 as a mediator of adipose tissue inflammation and insulin resistance. Diabetes 61, 615–625 (2012).
Shan, Y. X. et al. Hsp10 and Hsp60 modulate Bcl-2 family and mitochondria apoptosis signaling induced by doxorubicin in cardiac muscle cells. J. Mol. Cell. Cardiol. 35, 1135–1143 (2003).
Ghosh, J. C., Dohi, T., Kang, B. H. & Altieri, D. C. Hsp60 regulation of tumor cell apoptosis. J. Biol. Chem. 283, 5188–5194 (2008).
Chaiwatanasirikul, K. A. & Sala, A. The tumour-suppressive function of CLU is explained by its localisation and interaction with HSP60. Cell Death Dis. 2, e219 (2011).
Gupta, S. C. et al. Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxid. Redox Signal. 16, 1295–1322 (2012).
Tang, D. et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell. Metab. 13, 701–711 (2011).
Tatsumi, T. et al. Intracellular ATP is required for mitochondrial apoptotic pathways in isolated hypoxic rat cardiac myocytes. Cardiovasc. Res. 59, 428–440 (2003).
Yin, J. et al. Release and EGF receptor activation in epithelial cells. J. Cell. Sci. 120, 815–825 (2007). (Pt 5).
Abas, L., Bogoyevitch, M. A. & Guppy, M. Mitochondrial ATP production is necessary for activation of the extracellular-signal-regulated kinases during ischaemia/reperfusion in rat myocyte-derived H9c2 cells. Biochem. J. 349, 119–126 (2000).
Gantois, I. et al. Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nat. Med. 23, 674–677 (2017).
Zhu, J. et al. NF-kappaB p65 overexpression promotes bladder cancer cell migration via FBW7-mediated degradation of RhoGDIalpha protein. Neoplasia 19, 672–683 (2017).
Wang, C. J. et al. Survivin expression quantified by Image Pro-Plus compared with visual assessment. Appl. Immunohistochem. Mol. Morphol. 17, 530–535 (2009).
Fernandez-Vizarra, E. et al. Isolation of mitochondria for biogenetical studies: an update. Mitochondrion 10, 253–262 (2010).
Fang, H. et al. Mitochondrial DNA haplogroups modify the risk of osteoarthritis by altering mitochondrial function and intracellular mitochondrial signals. Biochim. Biophys. Acta 1862, 829–836 (2016).
Chen, T. H. et al. Knockdown of Hspa9, a del(5q31.2) gene, results in a decrease in hematopoietic progenitors in mice. Blood 117, 1530–1539 (2012).
Huang, H. et al. RhoGDIbeta promotes Sp1/MMP-2 expression and bladder cancer invasion through perturbing miR-200c-targeted JNK2 protein translation. Mol. Oncol. 11, 1579–1594 (2017).
Gong, S. et al. A deafness-associated tRNAHis mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res. 42, 8039–8048 (2014).
Acknowledgements
We thank the members in Jianxin Lyu’s laboratory for valuable discussions on this work. This work was supported by grants from the Chinese National Science Foundation (31670784 and 31671486), Zhejiang Provincial Natural Science Foundation of China (LY15H060007), and Foundation from Science and Technology Program of Wenzhou (Y20150045).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, C., Sun, H., Zheng, C. et al. Oncogenic HSP60 regulates mitochondrial oxidative phosphorylation to support Erk1/2 activation during pancreatic cancer cell growth. Cell Death Dis 9, 161 (2018). https://doi.org/10.1038/s41419-017-0196-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-017-0196-z
This article is cited by
-
Cysteine protease inhibitor 1 promotes metastasis by mediating an oxidative phosphorylation/MEK/ERK axis in esophageal squamous carcinoma cancer
Scientific Reports (2024)
-
Deoxycytidine kinase inactivation enhances gemcitabine resistance and sensitizes mitochondrial metabolism interference in pancreatic cancer
Cell Death & Disease (2024)
-
Antibody-modified integrated microfluidic terahertz biosensor for detection of breast cancer biomarkers
Optoelectronics Letters (2024)
-
Knockdown of heat shock protein family D member 1 (HSPD1) promotes proliferation and migration of ovarian cancer cells via disrupting the stability of mitochondrial 3-oxoacyl-ACP synthase (OXSM)
Journal of Ovarian Research (2023)
-
Biallelic FBXW7 knockout induces AKAP8-mediated DNA damage in neighbouring wildtype cells
Cell Death Discovery (2023)