Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus is highly contagious and causes lymphocytopenia, but the underlying mechanisms are poorly understood. We demonstrate here that heterotypic cell-in-cell structures with lymphocytes inside multinucleate syncytia are prevalent in the lung tissues of coronavirus disease 2019 (COVID-19) patients. These unique cellular structures are a direct result of SARS-CoV-2 infection, as the expression of the SARS-CoV-2 spike glycoprotein is sufficient to induce a rapid (~45.1 nm/s) membrane fusion to produce syncytium, which could readily internalize multiple lines of lymphocytes to form typical cell-in-cell structures, remarkably leading to the death of internalized cells. This membrane fusion is dictated by a bi-arginine motif within the polybasic S1/S2 cleavage site, which is frequently present in the surface glycoprotein of most highly contagious viruses. Moreover, candidate anti-viral drugs could efficiently inhibit spike glycoprotein processing, membrane fusion, and cell-in-cell formation. Together, we delineate a molecular and cellular rationale for SARS-CoV-2 pathogenesis and identify novel targets for COVID-19 therapy.
Similar content being viewed by others
Introduction
The current pandemic of coronavirus disease 2019 (COVID-19) is imposing tremendous threats to global public health. COVID-19 is caused by infection with the severe acute respiratory syndrome coronavirus 2 virus (SARS-CoV-2) [1,2,3], a member of the beta coronavirus clade that also contains SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), two well-known dangerous human viruses [4]. The SARS-CoV-2 virus is phylogenetically similar (~76% amino acid identity) to SARS-CoV [1,2,3], hence its name. In comparison with SARS-CoV and other members of the beta coronavirus clade, SARS-CoV-2 is much more contagious, particularly the G614 variant that is even more transmissible and infective than its D614 sister variant probably due to the bimodular effects of D614G mutation on the stability of spike trimer [5, 6], and the underlying mechanisms for this are of major academic and public interest. As of 26 Nov, 2020, more than sixteen millions of individuals worldwide were confirmed to have been infected with SARS-CoV-2, while only about tens of thousands of SARS-CoV and MERS-CoV patients were reported in total [7, 8]. The disease is clinically manifested as fever (>88%), respiratory symptoms (>67%), as well as lymphocytopenia (83%) [9]. Whereas the clinical features have been characterized in detail, the molecular and cellular mechanisms underlying COVID-19 pathogenesis remains largely elusive.
Cell-in-cell (CIC) structures, characterized by the presence of one or more viable cells inside of another cell, are a type of unique cellular structures that had been extensively documented in a wide range of human cancer tissues [10, 11], where the presence of cell-in-cell structures was demonstrated to regulate clonal selection [12] and genome stability [13, 14], and to profoundly impact patient outcomes as an independent prognostic factor [15,16,17]. Active intercellular interactions, controlled by a set of core elements [18,19,20], could produce CIC structures of homotypic (between same type of cells) and/or heterotypic (between different types of cells) [21, 22], both of which generally lead to the death of internalized cells in acidified vacuoles [10, 23]. Though the formation of heterotypic CIC structures were believed to constitute a mechanism of immune evasion in tumors by consuming functional immune cells, such as T lymphocytes or Nature killer cells [24, 25], its implications in the development of other diseases, such as the pathogenesis of infectious diseases, remains largely unknown.
Here, we demonstrated that the syncytia, resulted from SARS-CoV-2 infection, could target the infiltrated lymphocytes for internalization and CIC mediated death, contributing to lymphopenia in the patients with COVID-19. Moreover, a unique bi-arginine motif with the polybasic S1/S2 cleavage site of SARS-CoV-2 spike glycoprotein was identified to be capable of controlling this process by dictating membrane fusion and syncytia formation, which could be effectively inhibited by some candidate anti-viral drugs, such as arbidol. Our study uncovered a heretofore unrecognized mechanism underlying SARS-CoV-2 pathogenesis, and provided potentially novel targets for COVID-19 therapy.
Results
Prevalence of heterotypic cell-in-cell structures in COVID-19 lung autopsies
By examining a collection of lung tissue sections from six COVID-19 autopsies, we found that syncytia, a type of large cells with multiple nuclei that are negative in intercellular junction molecules such as E-cadherin and ZO-1 (Fig. 1a and Supplementary Fig. S1), were present in 10, and prevalent in 9 COVID-19 lung tissues, whereas the normal lung structures were completely disrupted (Fig. 1a), which was consistent with previous reports on COVID-19 autopsies [26,27,28,29]. Intriguingly, in addition to being positive for the SARS-CoV-2 spike (S) glycoprotein (Fig. 1a), a considerable number of syncytia were also found to enclose cells positive for CD45, a surface protein expressed by lymphocytes (Fig. 1a–c). These unique structures morphologically resemble the heterotypic CIC structures formed between lymphocytes and tumor cells, a pathological phenomenon generally documented in a wide range of human tumors as a potential mechanism of immune evasion by eliminating lymphocytes [22, 24]. Actually, more than three quarters of syncytia contained CD45-positive cells in all four lung tissue sections (Supplementary Fig. S1c), and each syncytium had ~3–4 CD45-positive cells on an average (Supplementary Fig. S1d, e), which were not detected in mononucleate non-syncytium cells. Moreover, the amounts of both syncytia, and syncytia containing CD45-positive cells as well, were inversely correlated with the amounts of lymphocytes in patients’ peripheral blood (Fig. 1d–f), suggesting that syncytia tend to internalize lymphocytes, conceivably contributing to lymphocyte loss in patients with COVID-19 [9].
a Representative images of a syncytium with CD45-positive cell internalized (indicated by white arrow) in a human COVID-19 lung specimen. White dashed lines depict shape of target syncytium. Tissue was stained with antibodies against SARS-CoV-2 S protein in green, E-cadherin in red and CD45 in magenta. Scale bars: 50 μm for the left image, 20 μm for zoomed images in the middle and multi-channel images on the right. Quantification of syncytia formation (b), and syncytia internalizing CD45-positive cells (c) in the lung specimens of patients with COVID-19. In total, more than 16 fields of view with a 20x objective lens per specimen were analyzed. d The percentage of lymphocytes in peripherial blood of COVID-19 patients during hospitalization. The peripherial lymphocytes were negatively associated with syncytia number (e) and cell-in-cell structures with CD45+ cells internalized by syncytia (f) in the lung tissues of patients with COVID-19. The purple liner trendlines were predicted based on data from all examined samples (n = 14); the red liner trendlines were predicted based on data from samples with cell-in-cell structures (CIC+) (n = 10).
Expression of the SARS-CoV-2 S glycoprotein induces syncytia
In order to establish a causal link between SARS-CoV-2 infection and syncytium formation, SARS-CoV-2 viruses were employed to infect Vero-E6 cells, a cell line used for routine virus culture. As expected, multinucleate giant syncytia were readily detected in S glycoprotein-positive cells 24 h post virus infection (Fig. 2a and Supplementary Fig. S2a). The S glycoprotein is the determinant for host entry of coronaviruses, and can be processed into an N terminal S1 fragment that is responsible for cellular receptor binding, and a C terminal S2 fragment that functions to promote membrane fusion [4]. To narrow down the effect of SARS-CoV-2 infection on membrane fusion, the S glycoprotein was expressed in 293 T cells stably expressing ACE2 (293T-ACE2), the cellular receptor for the S glycoprotein, and a number of large cells containing multiple nuclei (up to >60) were readily observed 12 h post transfection (Fig. 2b). To track the fusion process, time-lapse imaging was performed on 293T-ACE2 cells co-expressing S glycoprotein and Lyn-EGFP to label cell membranes. As a result, frequent fusion events were observed between neighboring cells as indicated by the disappearance of Lyn-EGFP-labeled membrane (Fig. 2c, Supplementary Movie S1), which eventually gave rise to multinucleate syncytium (Fig. 2a, b). During the fusion process, few fusion events were found to be taking place upon cell internalization, suggesting that SARS-CoV-2 S glycoprotein could initiate membrane fusion independent of the endocytosis process that is generally essential for host entry of other viruses via activation of pro-fusion proteins [30]. Rather, receptor binding to the cell surface is sufficient for the SARS-CoV-2 S glycoprotein to initiate membrane fusion (Supplementary Fig. S2d). Moreover, this S-mediated fusion occurred quite rapidly, with most fusions finishing within 10 min (Fig. 2d, e) and having an average fusion speed of 45.1 nm/s, ranging from 9.7 nm/s to 108.7 nm/s (Fig. 2f). The effect of S glycoprotein in inducing syncytia was confirmed in Hela cells expressing ACE2 as well (Supplementary Fig. S2b, c). Together, these data indicate that the SARS-CoV-2 infection can result in a rapid and endocytosis-independent membrane fusion via the expression of the S glycoprotein.
a Representative images of syncytia formation in Vero-ACE2 cells upon SARS-CoV-2 infection. Cells were stained with anti-SARS-CoV-2 S antibody in green. Control: no infection. Scale bars: 50 μm for the inserts in the lower left corner; 200 μm for full images. b Representative images of a syncytium formed in 293T-ACE2 cells expressing exogenous SARS-CoV-2 spike glycoprotein, Lyn-EGFP (cell membrane, green), and H2B-mCherry (nucleus, red). Scale bars: 20 μm. c Image sequence showing dynamic membrane fusion, indicated by the disappearance of Lyn-EGFP signal, in 293T-ACE2 cells expressing exogenous SARS-CoV-2 spike protein. Yellow arrows indicate sites where fusion is taking place. Scale bar: 20 μm. Related to Supplementary Movie S1. d Timeline graph showing representative fusion events. Each blue line indicates one fusion event. Blue circles indicate the beginning of membrane fusion. Red stars indicate the completion of membrane fusion. Quantification of the duration (e) and the speed (f) of membrane fusion induced by SARS-CoV-2 spike protein expression.
Syncytia target lymphocytes for cell-in-cell mediated death
We next examined whether the multinucleate syncytia produced by membrane fusion could internalize lymphocytes to form heterotypic cell-in-cell structures, CCRF-CEM, a human lymphoblast cell line isolated from a child with acute T cell leukemia, was engineered to stably express mCherry and co-cultured with 293T-ACE2 cells transfected with SARS-CoV-2 S glycoprotein. Cytospin analysis of this co-culture indicated that the syncytia could readily internalize multiple CCRF-CEM cells (Fig. 3a), as opposed to mononucleate 293T-ACE2 cells (Supplementary Fig. 3Sa, b). Syncytia with more nuclei or those that were larger in size were more likely to internalize CCRF-CEM cells (Fig. 3b, c and Supplementary S3c–h), and the number of internalized CCRF-CEM cells was positively correlated with the nucleus number in syncytia (Fig. 3d). In addition to CCRF-CEM, these syncytia could also efficiently internalize multiple types of leukocytes, including THP-1 cells, a monocyte cell line from an acute monocytic leukemia patient; Jurkat cells, a T lymphocytes cell line from an acute T cell leukemia patient; Raji cells, a B lymphocyte cell line from a Burkitt’s lymphoma patient; K562 cells, a human erythroleukemia line from a chronic myelogenous leukemia patient; as well as peripheral blood mononuclear cells (PBMC) isolated from a healthy man (Fig. 3e, f). Notably, by a time-lapse microscopy over 8 h, it was found that, except for those cells that stayed inside with no obvious changes or those with a minority of cell division, the internalized cells readily underwent cell death manifested by the progressive loss of Lyn-GFP, a cellular membrane marker (green arrow), as well as shrinkage and wrinkling of the cell body (white arrow), which was followed by corpse degradation as indicated by the diffusion of mCherry out of the dying cells (red arrow) (Fig. 3g, Supplementary Movie S2). The death of internalized CCRF-CEM cells took place within an average period of 182 min, ranging from 24 min to 429 min (Fig. 3h). Meanwhile, some cells, such as Raji cells and PBMCs, dies faster and in a higher percentage than others (Fig. 3h, i). The syncytia-mediated death turned out to be an efficient way to eliminate PBMC in a coculture experiment, where the amount of PBMC progressively decreased when cocultured with 293T-ACE2 cell expressing spike protein (293T-ACE2 spike) leading to syncytia formation; and by 24 h of coculture, more than 30% PBMC were cleared in 293T-ACE2 spike coculture as compared with those in control coculture (293T-ACE2 vector) (Fig. 3j, k and Supplementary S3i), which was positively correlated with syncytia formation (Fig. 3l). And the internalized PBMC were mostly positive in CD8, and CD4 to a less extend (Supplementary Fig. S4a, b), the pattern was confirmed in patient sample as well (Supplementary Fig. S4c). These results are consistent with an efficient clearance of lymphocytes, preferentially CD8+ T cells, by syncytia. To test whether the syncytia-mediated CIC formation was a potential target for anti-viral therapy, we examined the effects of candidate anti-viral compounds on these CIC structures using time-lapse microscopy. Six compounds were examined, including inhibitors of furin protease (6-D-Arg), cathepsin B/L (E64D), lysosome acidification (Con A, NH4Cl), and membrane fusion (Arbidol), and we also tested the compound hydroxychloroquine (HCQ). As shown in Fig. 3m, except for E64D, which displayed mild effects, these compounds profoundly inhibited the internalization of Raji cells by S glycoprotein-induced syncytia, a finding we also confirmed in PBMC cells (Fig. 3n). Together, these data suggest that syncytia could efficiently internalize lymphocytes for CIC mediated death, potentially contributing to lymphocyte loss in patients of COVID-19, and this could be prevented by treatment with some anti-viral compounds as a COVID-19 therapy.
a Representative images of a syncytium internalizing CCRF-mCherry cells to form cell-in-cell structures. Cells were stained with Phalloidin (green) and DAPI (blue). Scale bar: 20 μm. Graph plots of nuclei number (b) and area (c) of syncytia that internalizing CCRF-mCherry cells (CIC+, n = 41) or not (CIC−, n = 75). d Positive correlation between the nucleus number of syncytia and internalized CCRF-mCherry number. Analysis was performed by Spearman rank correlation. n = 41. Quantification of the formation frequency (e) and internalized cells (f) in cell-in-cell structures formed between syncytia and indicated cells. Data represent the mean ± SD of 10 or more fields with more than 100 syncytia analyzed each for e. n (left to right) = 44, 51, 28, 36, 43 and 45, respectively, for f. g Representative images and image sequence for the death of an internalized CCRF-mCherry cell within a syncytium. Green arrow indicates the dying of the internalized CCRF-mCherry cell; red arrow indicates the degradation of the internalized CCRF-mCherry cell. Scale bars: 20 μm for the left images; 10 μm for the right images. Plots of the duration (h) and frequency (i) of the death of the indicated cells in syncytia. n (left to right) = 24, 28, 21, 55, 24 and 38, respectively, for h. Data are the mean ± SD from 10 or more fields with more than 50 cell-in-cell structures analyzed each for i. j Representative FCM graphs for the respective cocultures of PBMC with 293T-ACE2-vector and 293T-ACE2-spike cells for different periods as indicated. k The quantification of PBMC changes over the indicated times in co-culture experiments. j The right Y axis is for the PBMC ratio between the two co-culture experiments (293T-ACE2-vector and 293T-ACE2-spike, respectively). Data are the mean ± SD of results from triplicate experiments. **p < 0.05, **p < 0.001, ***p < 0.0001. PBMC were added into the 293T-ACE2 cells 12 h post transfection with the empry vector or spike construct, respectively. l Quantification of syncytia formation in 293T-ACE2-spike cells at the indicated time points post PBMC adding. Data are the mean ± SD of results from 5 fields (20x objective lens) each. Note: syncytia were not formed in 293T-ACE2-vector cells. The inhibitory effects of the indicated compounds on the formation of cell-in-cell structures formed between syncytia and Raji cells (m), or PBMC (n). Data are the mean ± SD from 10 or more fields with more than 100 syncytia analyzed for each field. **p < 0.001, ***p < 0.0001.
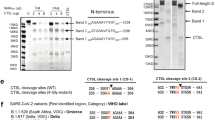
Pivotal role of the pre-cleavage amino acid composition in syncytium formation
Interestingly, we found that, in contrast to the S glycoprotein of SARS-CoV-2, the expression of S glycoprotein by the SARS-CoV virus, a genetically adjacent coronavirus that causes SARS in humans, could not induce membrane fusion and syncytia in 293T-ACE2 cells to any appreciable degree (Supplementary Fig. S5a). This was unlikely due to their differences in the S2 region, which is responsible for membrane fusion, as the SARS-CoV S2 region was highly similar (>96% similar, ~90% identical in amino acids) to that of SARS-CoV-2 (Supplementary Fig. S6) and could effectively mediate membrane fusion upon trypsin treatment (Supplementary Fig. S5b). Thus, it was likely that a “switch-on” element for membrane fusion was embedded into the S glycoprotein of SARS-CoV-2 that was not present in the SARS-CoV S glycoprotein. To identify the putative “switch-on” element, we first compared the amino acid sequences of their respective S glycoproteins. There were four major insertions in the S glycoprotein of SARS-CoV-2 as compared with that of SARS-CoV (Supplementary Fig. S6). Three of them were within the S1 fragment, a domain responsible for receptor binding, and are shared by the closely related coronaviruses from bat (RaTG13) or pangolin (PanCoV). In contrast, the fourth insertion of four amino acids (PRRA) was just before the S1/S2 cleavage site (Fig. 4a), and seemed to be specific for SARS-CoV-2 (Fig. 4a and Supplementary Fig. S6). We therefore hypothesized that the “PRRA” insertion might be the switch for S-mediated membrane fusion of SARS-CoV-2. To test this idea, a truncated mutant with “PRRA” deletion (2 ∆PRRA), analogous to the S glycoproteins from in RaTG13 or PanCoV, was constructed (Fig. 4b) and expressed in 293T-ACE2 cells. In agreement with our hypothesis, the “2 ∆PRRA” mutant completely lost the ability to induce syncytia (Fig. 4c, d), suggesting that the “PRRA” was a gain of function insertion in terms of membrane fusion. To further test this idea, the “VSLL” of the SARS-CoV S glycoprotein was replaced with a pre-cleavage motif of eight amino acids (QTNSPRRA), predicted using the PSI-BLAST algorithm (Supplementary Fig. S7a), to produce a SARS-CoV S mutant with “PRRA” (wPRRA) (Fig. 4b). The “wPRRA” mutant could induce syncytia as efficiently as SARS-CoV-2 S did, for which the “PRRA” insertion was also essential as depletion of the “PRRA” effectively abolished the acquired pro-fusion function of the SARS-CoV S “wPRRA” mutant (Fig. 4c, d). Cross-replacing the pre-cleavage motif (QTNSPPRA) in SARS-CoV-2 S with “VSLL” from SARS-CoV S also resulted in the inability to induce syncytia. Remarkably, the pro-fusion phenotype was tightly correlated with the production of the S2 fragment, the executor of the membrane fusion [4], as detected by an S2-specific antibody. Thus, these data support a gain of function role for the “PRRA” insertion in switching on membrane fusion.
a Alignment of spike protein sequences flanking the S1/S2 cleavage site from SARS-CoV (1SARS-S), pangolin coronavirus (3PanCOV), bat coronavirus (2RaTG13), SARS-CoV-2. Arrow indicates the S1/S2 cleavage site. b The pre-cleavage sequences (in blue) for the indicated mutants. “RS” in red indicates the S1/S2 cleavage site. c Representative cropped images for cell fusion upon expression of the indicated mutants. Scale bar: 100 μm. Quantification of syncytia formation (d) upon expression of the indicated mutants as detected by western blot (e). Data are the mean ± SD of results from 4-5 fields (20x objective lens) each for d. f SARS-CoV-2 spike mutants with indicated single or combined residue replacement with “A” in red, or the sequences of the pre-cleavage motif from surface glycoprotein of H1N1 or H7N1. Arrow indicates the S1/S2 cleavage site. Green shadow indicates two critical arginine for cleavage. Quantification of the syncytia formation (g) upon expression of the indicated mutants as detected by western blot (h). Data are the mean ± SD of results from 4-5 fields (20x objective lens) each for g. i 3-dimensional structure modeling of the SARS-CoV-2 spike monomer colored by the secondary structure. The zoomed images indicate the spatial residue patterns (0–4 positions shown in f), in the style of scaled ball and stick, for the cleavage sites of SARS-CoV-2 spike (2S), or H7N1 mutant (H7). Amino acid residues were indicated in number for the upper image (2S). S1 RBD: receptor binding domain in S1 fragment; S1 NTD: N terminal domain in S1 fragment. Effects of the indicated compounds on the processing of the spike protein (j) and syncytia formation (k). Data are the mean ± SD of results from 4-5 fields (20x objective lens) each for k.
A bi-arginine motif in spike glycoprotein dictates syncytium formation
Furthermore, to map the amino acids responsible for the aforementioned phenotypes, we made SARS-CoV-2 S mutants with single or combined mutations in the “PRRA” insertion, as well as the following amino acids (R), just before the S1/S2 cleavage site as depicted in Fig. 4f. While the P681A and R683A mutants could still effectively induce syncytia, mutants containing either R682A, or R685A, or both, failed to induce syncytia anymore (Fig. 4g), which was correlated with inhibited S2 production (Fig. 4h), suggesting that R682 and R685, but not R683, were required for S glycoprotein cleavage and membrane fusion. Consistent with this, by modeling the 3D structure of the S1/S2 cleavage site of SARS-CoV-2 S (Supplementary Fig. S7b and d), we observed that the side chains of both R682 and R685 were on the protein surface, which would conceivably facilitate interactions with proteases for efficient cleavage (Fig. 4i). On the contrary, the R683 residue was embedded in the protein and less likely to be reached (Fig. 4i and Supplementary Fig. S7d). Interestingly, a similar bi-arginine motif (RxxR), present in the SARS-CoV-2 S glycoprotein, was also identified in the pre-cleavage site of surface glycoproteins of other highly contagious viruses, such as the RSV, HIV, and H5N3 and H7N1 flu viruses, but not in other species of flu viruses that are less contagious such as H1N1 (Supplementary Fig. S8). Thus, the bi-arginine motif might be a conserved molecular basis for virus contagiousness by mediating surface glycoprotein cleavage. In agreement with this notion, transplanting the pre-cleavage motif of H7N1 surface glycoprotein into the SARS-CoV-2 S glycoprotein could also mediate S2 production and syncytium formation, as opposed to the low contagious H1N1 virus motif (Fig. 4f, h). In actuality, the side chain distribution pattern of the H7N1 pre-cleavage motif largely resembled that of the SARS-CoV-2 S glycoprotein very much (Fig. 4i), while the H1N1 pre-cleavage motif resembled those of the RR*2A and PRR*3A mutants in their spatial side chain distributions (Supplementary Fig. S7d). Moreover, some of the tested candidate anti-viral compounds, including 6-D-Arg, Con A, NH4Cl, Arbidol and hydroxychloroquine (HCQ), could efficiently inhibited S2 production and syncytium formation as well (Fig. 4j, k).
Discussion
In summary, we found that SARS-CoV-2 infection resulted in the production of multinucleate syncytia, which could target lymphocytes for internalization and cell-in-cell mediated elimination, potentially contributing to lymphopenia and pathogenesis in COVID-19 patients. This was dictated by a “switch on” element comprised of a bi-arginine motif in the S glycoprotein of SARS-CoV-2, which controlled the production of the S2 fusion fragment (Fig. 5). Candidate anti-viral compounds could effectively block S2 production, membrane fusion, and lymphocyte internalization. These data suggest that blocking surface glycoprotein cleavage, by strategies such as targeting the bi-arginine motif, might serve as a potential strategy to alleviate the pathogenesis caused by SARS-CoV-2 and conceivably other highly contagious viruses that contain the bi-arginine motifs.
The infection of ACE2-expressing cells by SARS-CoV-2 virus leads to the surface expression of viral spike glycoprotein, which harbors a bi-arginine motif that is required for protease-mediated processing and controls membrane fusion. The engagement of spike protein with its receptor ACE2 triggers membrane fusion, mediated by the S2 domain of the viral spike glycoprotein, between the neighboring cells, leading to the production of multinucleated syncytium. The syncytia are capable of targeting lymphocytes for internalization and cell-in-cell mediated death, conceivably contributing to lymphopenia in COVID-19 patients.
It is noted that the insertion of the “PRRA” residues, together with the following arginine (R), was predicted to create a polybasic furin cleavage site with few experimental evidence [31]. We demonstrated here that 6-D-Arg, a furin inhibitor, could significantly, though not as completely as the other four tested inhibitors, suppress the processing of the S glycoprotein into S2 as well as inhibit membrane fusion (Fig. 4j, k), supporting a role of furin-mediated cleavage in SARS-CoV-2 infection. Meanwhile, though the benefits of hydroxychloroquine treatment in patients with COVID-19 remain debatable [32], its anti-SARS-CoV-2 activities have been firmly demonstrated despite the underlying mechanisms being largely speculative [33, 34]. In this study, we unexpectedly identified hydroxychloroquine as a potent inhibitor of the processing of the S glycoprotein and membrane fusion (Fig. 4j, k), as well as syncytia-mediated cell-in-cell formation (Fig. 3m, n), which provides a molecular and cellular rationale for hydroxychloroquine treatment of COVID-19. In actuality, two recent in silico studies have predicted that hydroxychloroquine may directly interfere with the binding of the SARS-CoV-2 S glycoprotein to its receptors [35, 36], which is in good agreement with our data that receptor engagement promotes the S glycoprotein processing (Fig. 4e) that can be effectively blocked by hydroxychloroquine (Fig. 4j). Further validation of this mechanism, which is beyond the scope of this study, may help with the development of novel anti-SARS-CoV-2 strategies.
Notably, during the production of this manuscript, Braga et al. reported in Nature the prevalence of syncytial pneumocytes with multiple nuclei in the post-mortem samples of 41 patients with COVID-19, which was ascribed to the fusogenic activity of the SARS-CoV-2 S glycoprotein. And the S glycoprotein was found to upregulate TMEM16F, a chloride channel and lipid scramblase, to promote membrane fusion. Targeted inhibition of TMEM16F by compounds such as Niclosamide could readily block S glycoprotein-induced membrane fusion and syncytium formation, which was implicated in the therapy of COVID-19 [37]. Their results echo well our findings in this study that S glycoprotein-induced syncytium formation potentially underlies the pathogenesis of COVID-19 and may serve as a therapeutic target.
Materials and methods
Cell culture and compounds
The 293 T, 293T-ACE2 and Hela-ACE2 cells were maintained in DMEM (MACGENE Tech Ltd., Beijing, China) supplemented with 10% fetal bovine serum (Kang Yuan Biol, Tianjin, China) and 1% Penicillin-Streptomycin (MACGENE Tech Ltd., Beijing, China). CCRF-CEM, Jurkat, K562, Raji, THP-1 and PBMC cells were cultured in RPMI-1640 (MACGENE Tech Ltd., Beijing, China) supplemented with 10% fetal bovine serum (Kang Yuan Biol, Tianjin, China). All cells were incubated with 5% CO2 at 37 °C. Detail information could be found in Supplementary Table S1 for cell lines and Supplementary Table S2 for compounds.
Constructs
The construct expressing SARS S protein in pSecTag2-Hygro-A was a gift from Dr. Cheng Cao (Beijing Institute of Biotechnology). The codon-optimized SARS-CoV-2 S cDNA was synthesized at Genscript Biotech Corporation (Nanjing, China). The wild type S genes of SARS-CoV and SARS-CoV-2 and their mutants were cloned into pSecTag2-Hygro-A through seamless homologous recombination. Detail information could be found in Supplementary Table S3 for constructs.
Cell fusion induced by SARS-CoV-2 spike protein
For cell fusion assay, about 6.5 × 105 cells were plated per well in 6-well plate precoated with type I collagen (354236, BD Bioscience) and cultured for 24 h. Cells were then transfected with respective constructs by Lipofectamine 2000 reagent (11668019, Thermal Fisher Scientific, US) following the protocol provided. Images of 5 fields (20x objective lens) were taken on Hoechst-stained cells 24 h post transfection by Nikon microscope. Nucleus counting was performed by NIS elements AR software (Nikon, Japan). The fusion index (FI) was calculated as “% of nuclei in fused cells”.
For time-lapse imaging of membrane fusion, 293T-ACE2 cells, plated on glass bottom plate (Cellvis.), were first transfected with constructs expressing Lyn-EGFP (labeling membrane), H2B-mCherry (labeling nucleus) and SARS-CoV-2 spike for 6 h, and then imaged on a Z-stack with Ultraview Vox confocal system (Perkin Elmer) on Nikon Ti-E microscope. Fluorescent and differential interference contrast images were captured every 5 min for 4 h. The length measurement was performed by the MEASUREMENTS module in Volocity software (V6.3, Perkin Elmer), the fusion speed was calculated by dividing the length of fused membrane with time duration of fusion.
PEG-induced cell fusion and cell-in-cell formation
About 1 × 107 tumor cells were collected and washed with serum-free DMEM. 500 μL of PEG 1450 (#P7181, sigma) were added to the cell pellet in a corning Tube followed by gently stirring for 2 min in a 37 °C water bath. Then 10 mL of DMEM were added to the cell suspension over the next 5 min with constant stirring and let stand for 1 min. Cells were collected at 400 g for 5 min to remove PEG, and washed with PBS for three times. Quantification of cellular DNA was carried out using Cell Cycle Detection Kit (#KGA511, Keygen). For cell-in-cell formation assay, the tumor cells resuspended with complete medium were seeded in 6-well plate for 12 h followed by coculturing with NK cells for 4 h before fixed for cell-in-cell quantification.
PBMC clearance assay
293T-ACE2 cells were transfected with construct expressing SARS-CoV-2 spike for 12 h to allow syncytium formation, then cocultured with PBMC cells prestained with 2.5 μM CellTracker Orange (Invitrogen, San Diego, CA, USA). Total cocultured cells were collected at 6, 12 and 24 h after the addition of PBMC, respectively, for flow cytometry analysis. Data were acquired on a NovoCyte flow cytometer (Agilent, USA) and analyzed using Novoexpress V1.4.1.software.
Western blotting
Cells were lysed on ice with cold RIPA buffer containing phosphatase-protease inhibitors (CWBiotech, Beijing) for 20 min followed by ultrasound (power 40%, work 6 s, stop 9 s, 5 times in total). After being centrifuged at 12,000 rpm for 10 min, the supernatant was collected for SDS-PAGE electrophoresis followed by transferring onto the PVDF membrane (0.2 μm, Millipore). The PVDF membrane, blocked with 5% skimmed milk for 1 h at room temperature, was then blotted with primary antibodies in 5% BSA for 12 h at 4 °C or 4 h at room temperature, followed by one-hour secondary antibodies at room temperature. The primary antibodies used: ACE2 (Proteintech, 1:3000, 66699-1-Ig), SARS-CoV/SARS-CoV2(COVID-19) spike (GeneTex, 1:2000, GTX632604), α-Tubulin (Proteintech, 1:1000, 11224-1-AP). The secondary antibodies used: anti-rabbit IgG HRP (CST, 1:3000, #7074), anti-mouse IgG HRP (CST, 1:3000, #7076). Detail information could be found in Supplementary Table S4 for antibodies.
Cell-in-cell formation assay
For cytospin analysis, mCherry labeled CCRF-CEM cells were cocultured for 8 h with 293T-ACE2 cells that had been transfected with SARS-CoV-2 S construct for 14 h. Subsequently, cytospins were prepared by Cytocentrifuge 7620 (Wescor, Logan, UT, USA) at 400 rpm for 3 min, and then fixed with 4% paraformaldehyde followed by staining with Alexa Fluor®488 Phalloidin (1:200; Life technololgies; A12379) and DAPI for 20 min. Cell-in-cell structures were examined by Nikon Ti-E microscope equipped with motorized stage and Neo Vacuum cooled Scientific CMOS Camera (Andor Technology), and quantified as “% of syncytia containing CCRF-CEM”.
For time-lapse analysis, 293T-ACE2 cells, plated on glass bottom plate (Cellvis), were first transfected with Lyn-EGFP (labeling membrane) and SARS-CoV-2 spike for 14 h to allow syncytium formation, then cocultured with CCRF-CEM-mCherry, or immune cell lines pre-stained with 2.5 μM CellTracker Orange (Invitrogen, San Diego, CA, USA), for 3 h to allow cell adhesion and early cell-in-cell formation. Wide field imaging was then performed by Nikon Ti-E microscope equipped with motorized stage and Neo Vacuum cooled Scientific CMOS Camera (Andor Technology). Images were collected every 10 min for 12 h using 20x Apo objective lens with 15 ms exposure for DIC channel, 100 ms exposure for mCherry channel and 150 ms exposure for FITC channel. Cells were cultured in humidified chamber supplied with 5% CO2 at 37°C during imaging. Image sequences were reviewed using Nikon NIS-Elements AR 4.5 software. For compound treatment, compounds were added together with immune cell lines to 293T-ACE2 cells prepared above. Please find in Supplementary Table S2 for detail information on the compounds used in this study.
3D time-lapse imaging was performed on cells prepared as above with Ultraview Vox confocal system (Perkin Elmer) on Nikon Ti-E microscope. Fluorescent and differential interference contrast images were captured every 10 min for 12 h.
SARS-CoV-2 virus infection
African green monkey kidney Vero E6 cell line stably expressing ACE2 (Vero-ACE2) was maintained in minimum Eagle’s medium (MEM; Gibco Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gibco Invitrogen) at 37 °C in a humidified atmosphere of 5% CO2. The 2019-CoV-2 (GenBank ID: MT627325), a clinical isolate of SARS-CoV-2 virus, was propagated in Vero E6 cells, and viral titer was determined by 50% tissue culture infective dose (TCID50) using immunofluorescence assay. All the infection experiments were performed in a biosafety level-3 (BLS-3) laboratory in the Department of Microbiology at the 2nd Military Medical University. For syncytium formation, Vero-ACE2 cells (2 × 104 cells/well) in 96-well Cell culture plate were first infected with 2019-Cov-2 (MOI of 0.1) for 24 h, and then cultured in normal medium overnight before preceding to immunofluorescence staining with anti-SARS-CoV-2 S1 Antibody (1:200, Sino Biological, #40150-R007) following standard protocol described previously [38]. The images were taken by fluorescence microscopy.
Patients specimens and ethics
The autopsy for COVID-19 death was carried out with informed consent under the approval of Ethics Committee of Wuhan Infectious Diseases Hospital (KY-2020-15.01) and Ethics Committee of the First Affiliated Hospital of Army Medical University (KY2020298), complying with regulations issued by the National Health Commission of China and the Helsinki Declaration, and in accordance with the National standards of Examination of Cadaver’s Surface in Forensic Medicine (GA/T 149-1996) and Autopsy in Forensic Medicine (GA/T 147-1996) of China. The dry autopsy, which didn’t produce any liquids during the whole process of autopsy to ensure biosafety, was employed to collect lung tissues in negative pressure operating room following the Methods of Collecting, Fixing, Packing and Sending for Forensic Pathological Material (GA/T 148-1996). The lung tissues were immersed immediately into 10% neutral buffered formalin for 7 days to kill the SARS-CoV-2 virus before proceeding to section preparation following the standard histological procedures. Sections of 4-5 μm were prepared for subsequent staining. Detail information on each specimen could be found in Supplementary Table S5 for specimens.
Immunostaining and image processing
For patient tissues, the “EML” multiplex staining method was used to co-stain multiple cell types as previously reported [22]. In brief, the deparaffined slides were first stained with primary antibody against CD45 (rabbit pAb from Proteintech, 20103-1-AP) at a dilution of 1:1500 using Opal Multiplex tissue staining kit (Perkin Elmer, NEL791001KT) according to the standard protocol, and the CD45 molecule was eventually labeled with Cyanine 5 fluorophore. Slides were then incubated with mixed primary antibodies against E-cadherin (mouse mAb from BD Biosciences, 610181), or ZO1 (mouse mAb from Proteintech, 66452-1-Ig), together with SARS-CoV Spike S1 Subunit (rabbit pAb from Sino biological, 40150-T52) which was a gift from Dr. Guan Yang (Beijing institute of Biotechnology), followed by incubating with secondary antibodies of Alexa Fluor 568 anti-mouse antibody (Invitrogen, A11031) and Alexa Fluor 488 anti-rabbit antibody (Invitrogen, A11034).
For cultured cells or cytospin, samples were fixed with 4% PFA and then permeabilized with 0.2% TritonX-100 before blocked with 5% BSA for 1 h at room temperature. Primary antibodies (anti-E-cadherin, 1:200, BD Biosciences, 610181; anti-ZO-1, 1:500, Proteintech, 66452-1-Ig; anti-CD4, 1:200, Abclonal, A19018; anti-CD8, 1:200, abcam, ab34364) were applied and incubated at 4 °C overnight, followed by one-hour incubation of secondary antibodies (Invitrogen, A11031 or A11034) after thoroughly washing with PBS of 3 times.
All slides were counterstained with DAPI to show nuclei and mounted with Prolong Gold antifade reagent (Invitrogen) and cover slips. Images were captured and processed by Ultraview Vox confocal system (Perkin Elmer) or Widefield Fluorescence system (Nikon, Japan) on Nikon Ti-E microscope. Detail information could be found in Supplementary Table S4 for antibodies and related reagents.
Bioinformatics
Sequence alignment was performed by the Clustal Omega software (http://www.clustal.org/) or Align algorithm at UniProt (https://www.uniprot.org/align/). The pre-cleavage motif was identified by Position-Specific Iterated BLAST (PSI-BLAST) algorithm queried with sequence “GAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNS” at the National Center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/). The 3D structure modeling of SARS-CoV-2 S glycoprotein was performed by the Modeling algorithm at SWISS-MODEL (https://swissmodel.expasy.org/) with the template of 6vxx.pdb from RSCB protein data bank (http://www.rcsb.org/).
Statistics
Data were expressed as means with standard deviations (SD). P values were calculated using two-tailed Student’s t test from Excel or GraphPad Prism software, and P values less than 0.05 were considered statistically significant. Spearman rank correlation analysis were performed to examine the correlation between factors of interest.
References
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3.
Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;https://doi.org/10.1038/s41586-020-2008-3.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl J Med. 2020;382:727–33.
Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–61.
Jiang X, Zhang Z, Wang C, Ren H, Gao L, Peng H, et al. Bimodular effects of D614G mutation on the spike glycoprotein of SARS-CoV-2 enhance protein processing, membrane fusion, and viral infectivity. Signal Transduct Target Ther. 2020;5:268–71.
Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–51.e8.
Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–3.
WHO. Coronavirus disease (COVID-2019) situation report-174. Situation reports. 2020;doi: 20200318-sitrep-174-covid-19:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020; https://doi.org/10.1056/NEJMoa2002032.
Huang H, Chen Z, Sun Q. Mammalian cell competitions, cell-in-cell phenomena and their biomedical implications. Curr Mol Med. 2015;15:852–60.
Mackay HL, Muller PAJ. Biological relevance of cell-in-cell in cancers. Biochem Soc Trans. 2019;47:725–32.
Sun Q, Luo T, Ren Y, Florey O, Shirasawa S, Sasazuki T, et al. Competition between human cells by entosis. Cell Res. 2014;24:1299–310.
Rizzotto D, Villunger A. P53 clears aneuploid cells by entosis. Cell Death Differ. 2020; https://doi.org/10.1038/s41418-020-00659-x.
Liang J, Niu Z, Zhang B, Yu X, Zheng Y, Wang C, et al. p53-dependent elimination of aneuploid mitotic offspring by entosis. Cell Death Differ. 2021;28:799–813.
Zhang X, Niu Z, Qin H, Fan J, Wang M, Zhang B, et al. Subtype-based prognostic analysis of cell-in-cell structures in early breast cancer. Front Oncol. 2019;9:895.
Huang H, He M, Zhang Y, Zhang B, Niu Z, Zheng Y, et al. Identification and validation of heterotypic cell-in-cell structure as an adverse prognostic predictor for young patients of resectable pancreatic ductal adenocarcinoma. Signal Transduct Target Ther. 2020;5:246–8.
Fan J, Fang Q, Yang Y, Cui M, Zhao M, Qi J, et al. Role of heterotypic neutrophil-in-tumor structure in the prognosis of patients with Buccal Mucosa squamous cell carcinoma. Front Oncol. 2020;10:541878.
Wang M, Niu Z, Qin H, Ruan B, Zheng Y, Ning X, et al. Mechanical ring interfaces between adherens junction and contractile actomyosin to coordinate entotic cell-in-cell formation. Cell Rep. 2020;32:108071.
Sun Q, Cibas ES, Huang H, Hodgson L, Overholtzer M. Induction of entosis by epithelial cadherin expression. Cell Res. 2014;24:1288–98.
Niu Z, He M, Sun Q. Molecular mechanisms underlying cell-in-cell formation: core machineries and beyond. J Mol Cell Biol. 2021; https://doi.org/10.1093/jmcb/mjab015.
Ruan B, Niu Z, Jiang X, Li Z, Tai Y, Huang H, et al. High frequency of cell-in-cell formation in heterogeneous human breast cancer tissue in a patient with poor prognosis: a case report and literature review. Front Oncol. 2019;9:1444.
Huang H, Chen A, Wang T, Wang M, Ning X, He M, et al. Detecting cell-in-cell structures in human tumor samples by E-cadherin/CD68/CD45 triple staining. Oncotarget. 2015;6:20278–87.
Su Y, Ren H, Tang M, Zheng Y, Zhang B, Wang C, et al. Role and dynamics of vacuolar pH during cell-in-cell mediated death. Cell Death Dis. 2021;12:119.
Fais S, Overholtzer M. Cell-in-cell phenomena in cancer. Nat Rev Cancer. 2018;18:758–66.
Wang S, He MF, Chen YH, Wang MY, Yu XM, Bai J, et al. Rapid reuptake of granzyme B leads to emperitosis: an apoptotic cell-in-cell death of immune killer cells inside tumor cells. Cell Death Dis. 2013;4:e856.
Luo W, Yu H, Gou J, Li X, Sun Y, Li J, et al. Clinical pathology of critical patient with novel coronavirus pneumonia (COVID-19). Preprint at https://www.preprints.org/manuscript/202002.0407/v1.
Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020; https://doi.org/10.1016/s2213-2600(20)30076-x.
Fox SE, Akmatbekov A, Harbert JL, Li G, Brown JQ, Vander Heide RS. Pulmonary and cardiac pathology in Covid-19: the first autopsy series from New Orleans. medRxiv. 2020; https://doi.org/10.1101/2020.04.06.20050575:2020.04.06.20050575.
Zhou L, Niu Z, Jiang X, Zhang Z, Zheng Y, Wang Z, et al. SARS-CoV-2 targets by the pscRNA profiling of ACE2. TMPRSS2 Furin Proteases iScience. 2020;23:1–15.
Boonstra S, Blijleven JS, Roos WH, Onck PR, van der Giessen E, van Oijen AM. Hemagglutinin-mediated membrane fusion: a biophysical perspective. Annu Rev Biophys. 2018;47:153–73.
Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–2.
Ledford H. Safety fears over drug hyped to treat the coronavirus spark global confusion. Nature 2020;582:18–9.
Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Disco. 2020;6:16.
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–71.
Fantini J, Di Scala C, Chahinian H, Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55:105960.
Amin M, Abbas G. Docking study of Chloroquine and Hydroxychloroquine interaction with SARS-CoV-2 spike glycoprotein-An in silico insight into the comparative efficacy of repurposing antiviral drugs. J Biomol Struct Dyn. 2020; https://doi.org/10.1080/07391102.2020.1775703:1-11.
Braga L, Ali H, Secco I, Chiavacci E, Neves G, Goldhill D, et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 Spike-induced syncytia. Nature. 2021; https://doi.org/10.1038/s41586-021-03491-6.
Wang C, Chen A, Ruan B, Niu Z, Su Y, Qin H, et al. PCDH7 inhibits the formation of homotypic cell-in-cell structure. Front Cell Dev Biol. 2020;8:1–12.
Acknowledgements
We thank Dr. Cheng Cao, Dr. Hui Zhong and Dr. Guan Yang at Beijing Institute of Biotechnology for reagents. Dr. Hongguang Ren and Dr. Junjie Yue at Beijing Institute of Biotechnology for assistance in bioinformatics.
Funding
This work was supported by the Key Science & Technology Project of Beijing Educational Committee and the Beijing Municipal Natural Science Foundation (KZ202110025029 to HH), Beijing Municipal Administration of Hospitals Incubating Program (PX2021033 to HH), the National Key Research & Development Program of China (2018YFA0900804 to YZ and ZC, 2019YFA0903801 to QS), and the National Natural Science Foundation of China (81872314 to ZC, 31970685 to QS).
Author information
Authors and Affiliations
Contributions
Concept and design: QS; Phenotype: QS and ZZ; Tissue staining, imaging and analysis: ZN and YQW; Human specimen and sections: XY, XB, DF, MP, YYW and LL; Virus induced fusion: HP and PZ; S protein induced fusion and imaging: ZZ and ZN; Cell-in-cell experiments: YZ, YS, YCZ, MH; Gene cloning: BZ; Expression detection: CW; Data analysis: YCZ, MT, WG, XJ, HR, LG; Bioinformatics: QS; Figures: QS, HH and YCZ; Data interpretation: QS, HH; Manuscript: QS, GM, HH, XW, HS; Funding: QS, HH, YZ and ZC. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics statement
The autopsy for COVID-19 death was carried out with informed consent under the approval of Ethics Committee of Wuhan Infectious Diseases Hospital (KY-2020-15.01) and Ethics Committee of the First Affiliated Hospital of Army Medical University (KY2020298).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Y. Shi
Supplementary information
41418_2021_782_MOESM4_ESM.tif
Fig. S4 Subtyping T lymphocytes internalized into syncytia, and effects of candidate compounds on CIC formation. Related to Figure 3.
Rights and permissions
About this article
Cite this article
Zhang, Z., Zheng, Y., Niu, Z. et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ 28, 2765–2777 (2021). https://doi.org/10.1038/s41418-021-00782-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00782-3
This article is cited by
-
A high-throughput screening system for SARS-CoV-2 entry inhibition, syncytia formation and cell toxicity
Biological Procedures Online (2023)
-
Ultrastructural study confirms the formation of single and heterotypic syncytial cells in bronchoalveolar fluids of COVID-19 patients
Virology Journal (2023)
-
SARS-CoV-2 and innate immunity: the good, the bad, and the “goldilocks”
Cellular & Molecular Immunology (2023)
-
Inflammatory cell death, PANoptosis, screen identifies host factors in coronavirus innate immune response as therapeutic targets
Communications Biology (2023)
-
Long-term effects of SARS-CoV-2 infection on human brain and memory
Cell Death Discovery (2023)