Abstract
Although the interaction between tumors and tumor-associated macrophages (TAMs) has been reported to facilitate the targeted drug resistance and progression of clear cell renal cell carcinoma (ccRCC), the related mechanisms remain unknown. Here, we report that SOX17 serves as a novel tumor suppressor in ccRCC and a positive regulatory loop, SOX17low/YAP/TEAD1/CCL5/CCR5/STAT3, facilitates the ccRCC-TAM interaction. SOX17 expression was commonly downregulated and negatively correlated with TAM infiltration in ccRCC specimens, and the integration of SOX17 and TAMs with the existing clinical indicators TNM stage or SSIGN score achieved better accuracy for predicting the prognosis of ccRCC patients. Mechanistically, SOX17 knockdown activated YAP signaling by promoting the transcription and nuclear distribution of YAP, which recruited TEAD1 to trigger CCL5 transcription. Then, CCL5 educated macrophages toward TAMs, which reciprocally enhanced ccRCC progression through CCL5/CCR5 and activated STAT3/SOX17low/YAP. However, SOX17 overexpression in ccRCC achieved the opposite effect. Thus, a positive regulatory loop, SOX17low/YAP/TEAD1/CCL5/CCR5/STAT3, was identified in the ccRCC-TAM interaction. Furthermore, targeting tumor-TAM interactions by blocking this positive regulatory network impaired the metastasis and targeted drug resistance of ccRCC in in vivo mouse models of lung metastasis and orthotopic ccRCC. These findings provide a new mechanism underlying the tumor-TAM interplay in ccRCC progression and present a potential target for inhibiting targeted drug resistance and metastasis in advanced ccRCC.
Similar content being viewed by others
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common histological subtype of renal cell carcinoma (RCC) and accounts for the majority of deaths from kidney cancer [1]. Although radical surgical operation is an option for early ccRCC patients and targeted drugs improve survival in patients with advanced or metastatic ccRCC, the prognosis of patients is still poor [2]. Therefore, elucidating the related mechanisms will help to develop effective treatments for ccRCC patients.
Sex-determining region Y (SRY)-box transcription factor 17 (SOX17) encodes a member of the SOX family of transcription factors involved in the establishment of the endoderm-biased stem cell state, early human development and germ cell programming, and the conversion of human fibroblasts into endothelial cells and erythroblasts [3,4,5]. Recently, SOX17 has been shown to serve as a tumor suppressor in various types of malignant tumors, including cervical cancer, cholangiocarcinoma, and esophageal cancer [6,7,8]. Moreover, SOX17 restrains the proliferation and progression of tumors by inactivating the Wnt/β-catenin pathway via upregulation of the levels of the Wnt antagonist secreted frizzled related protein-1 (SFRP1), trans-suppressing β-catenin expression, and enhancing the GSK3β-independent protein degradation of β-catenin. In addition, SOX17 suppresses β-catenin/T-cell factor-dependent transcription [9, 10]. In addition, low SOX17 expression in tumor specimens is a prognostic biomarker and predicts disease progression in patients [11]. However, the clinical significance and biological function of SOX17 in ccRCC have not been elucidated.
In addition to the intratumoral signaling pathway, the tumor microenvironment has been reported to sustain the survival of tumor cells [12, 13]. Monocytes and macrophages are major components of the tumor microenvironment and can be educated by tumor cells to transition into tumor-associated macrophages (TAMs) [14].
The infiltrating macrophages adapt a range of polarization states that can be classified into the M1 and M2 polarization models. M1 macrophages regulate defense of the host from external stimuli, while M2 macrophages facilitate tumor growth. TAMs are predominantly polarized toward a protumoral M2 phenotype in the tumor microenvironment [15]. Many studies, including ours, have reported that tumor-TAM interactions promote the progression of tumors [16,17,18]. In fact, ccRCC is frequently infiltrated with TAMs that can promote ccRCC progression [19] The interleukin-1 (IL-1)/IL-1 receptor axis facilitates the protumor phenotype of TAMs [20]. In addition, the intratumoral balance of diametrically polarized TAMs (CD11c+ M1 and CD206+ M2) is a novel independent predictor of survival in patients with ccRCC [15]. Therefore, eliminating the tumor-TAM network is important for gaining a good response to targeted drugs in ccRCC.
In this study, we demonstrated that SOX17 served as a prognostic indicator and a tumor suppressor in ccRCC, and a positive regulatory loop was identified to facilitate the interaction between ccRCC cells and TAMs.
Results
SOX17 is downregulated in ccRCC tissues and predicts a poor postoperative ccRCC patient prognosis
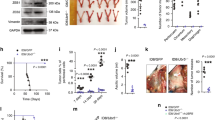
First, as shown in Fig. 1a, b and Fig. S1a, both mRNA and protein expression levels of SOX17 were observed to be lower in ccRCC specimens than in paired normal tissues. In addition, SOX17 expression in various ccRCC cell lines was downregulated compared with that in the normal renal cell line HK-2 (Fig. S1b-c). Moreover, immunohistochemistry (IHC) was performed in postoperative ccRCC specimens (n = 539), which confirmed that SOX17 expression was commonly downregulated in ccRCC (Fig. 1c).
a Real-time PCR assays were used to test the mRNA expression of SOX17 in ccRCC samples and paired adjacent renal tissues (n = 64). b Western blot assays were applied to examine the protein expression of SOX17 in ccRCC and peri-tumors (n = 30). c Representative images of hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) staining for SOX17 in the matched ccRCC and adjacent renal tissues are presented (n = 539; different scale bars are presented on the images). The expression of SOX17 was quantificationally determined by H-score. d Representative images of H&E and IHC staining of SOX17 expression in ccRCC specimens with different H-scores are shown (different scale bars are presented on the images). e–h Kaplan–Meier analyses for OS and PFS were performed in ccRCC patients from cohort 1 (n = 343) or cohort 2 (n = 115).
Next, an IHC assay demonstrated that lower SOX17 expression was observed in ccRCC with high TNM stages (Fig. S1d). Second, IHC was employed on ccRCC specimens from two independent cohorts of ccRCC patients (n = 458) to determine the clinical value of SOX17 (Fig. 1d). Then, the ccRCC patients in cohort 1 were divided into SOX17high and SOX17low groups, and the SOX17low group had higher World Health Organization/International Society of Urological Pathology grading (WHO/ISUP grading), TNM stages, and stage, size, grade, and necrosis (SSIGN) scores (Fig. S1e; Table S1). Furthermore, Kaplan–Meier survival analysis indicated that the SOX17low group presented worse OS and progression-free survival (PFS) than the SOX17high group (Fig. 1e, f). These results were also confirmed in cohort 2 (Fig. 1g, h; Table S1). Moreover, both cohorts of ccRCC patients were merged and randomly divided into two cohorts at a 3:2 ratio. Consistent with the above results, low SOX17 expression also predicted unfavorable clinicopathological characteristics and poor prognosis in ccRCC patients in the randomized training, validation, and combined cohorts (Fig. S1f–k; Table S1). In summary, low SOX17 expression is a helpful indicator for predicting ccRCC patients’ poor prognosis.
SOX17 exerts a tumor-suppressive role in ccRCC
We next examined the biological function of SOX17 in ccRCC cells. First, shRNAs targeting SOX17 were employed in the 786-O and 769-P ccRCC cell lines (Fig. S2a). CCK-8 and flow cytometry assays showed that compared with their respective control cells, ccRCC cells with SOX17 knockdown presented with increased proliferation and decreased apoptosis (Fig. 2a, b). Moreover, under treatment with the first-line targeted drugs pazopanib or sunitinib [2], compared with the control cells, 786-O cells with SOX17 knockdown exhibited increased proliferation and decreased apoptosis (Fig. 2c, d; Fig. S2b, c). But SOX17 knockdown did not regulate cell cycle of ccRCC cells (Fig. S2d–e). Moreover, the promoting effects of SOX17 knockdown on the invasion and migration abilities of ccRCC cells were also observed (Fig. 2e, f). Furthermore, compared with that of control cells, subcutaneous injection of 786-O cells with SOX17 knockdown resulted in increased volume and tumor growth of xenografts (Fig. 2g, h; Fig. S2f).
a A CCK-8 assay was used to detect the proliferation of 786-O or 769-P cells without or with SOX17 knockdown. b Flow cytometry assays were employed to test the percentage of apoptotic cells in 786-O or 769-P cells without or with SOX17 knockdown. 786-O cells without and with SOX17 knockdown were treated with pazopanib (6 μM). Cell proliferation was detected by CCK-8 assays for different times (c) and cell apoptosis was evaluated by flow cytometry assays (d). Representative images and statistical analysis on the results of the invasion (e) or migration (f) assays in 786-O or 769-P cells without or with SOX17 knockdown are shown (scale bar = 200 µm). g, h SOX17-knockdown or control 786-O cells were subcutaneously injected into nude mice (n = 5/group). The tumor xenografts derived from the two groups are shown (g). The volumes of the tumor xenografts from these two groups were compared at the indicated times (once a week for 6 weeks after inoculation) (h). i A CCK-8 assay was performed to test the proliferation of 786-O or 769-P cells without or with SOX17 overexpression. j Flow cytometry assays were performed to detect the percentage of apoptotic 786-O or 769-P cells without or with SOX17 overexpression. k, l 786-O cells without and with SOX17 overexpression were treated with pazopanib (6 μM). Cell proliferation was detected by CCK-8 assays for different times (k) and cell apoptosis was examined by flow cytometry (l). Representative images and statistical analysis of the invasion (m) or migration (n) assays with 786-O or 769-P cells without or with SOX17 overexpression are presented (scale bar = 200 µm). o, p SOX17-overexpressing or control 786-O cells were subcutaneously injected into nude mice (n = 5/group). The tumor xenografts derived from the two groups are shown (o). The volumes of the tumor xenografts from these two groups were compared at the indicated times (once a week for 6 weeks after inoculation) (p). All p values are defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Moreover, SOX17 overexpression decreased proliferation, induced more apoptosis, and inhibited the pazopanib or sunitinib resistance and the invasion and migration of ccRCC cells (Fig. 2i–n; Fig. S2g–i). In addition, SOX17 overexpression did not regulate cell cycle of ccRCC cells (Fig. S2j–k). Furthermore, SOX17 overexpression inhibited the growth of subcutaneous ccRCC xenografts (Fig. 2o, p; Fig. S2l). Therefore, SOX17 is a tumor-suppressor gene in ccRCC.
SOX17 restrains the growth and progression of ccRCC by inhibiting the nuclear distribution and transcription of YAP
To explore the related molecular mechanisms, RNA-sequencing was performed in 786-O cells without or with SOX17 knockdown, which identified the differentially expressed genes and pathways regulated by SOX17 (Fig. 3a, b, Fig. S3a, b, and Table S2). Among these genes and pathways, differential regulation of the Hippo pathway and higher expression of YAP (Fig. 3a, b; Table S2), which is the effector of the Hippo pathway and serves as an oncogene in various types of cancers [21], were observed in 786-O cells with SOX17 knockdown in contrast to the control 786-O cells. Then, real-time PCR assays validated that the expression levels of YAP and its target genes were increased in ccRCC cells with SOX17 knockdown, while SOX17 overexpression exhibited the opposite effects (Fig. 3c, d; Fig. S3c, d). YAP is phosphorylated and negatively regulated by the upstream kinase large tumor suppressor 1/2 (LATS1/2), and p-YAP is retained in the cytoplasm, which leads to less YAP entering the nucleus and thus does not trigger protumoral downstream genes [22]. Then, a nucleoplasm separation-based Western blot assay was employed and demonstrated that SOX17 knockdown decreased p-YAP in the cytoplasm and increased YAP expression in the nucleus of ccRCC cells, which was also confirmed by cellular immunofluorescence assays (Fig. 3e; Fig. S3e, f). Moreover, overexpressing SOX17 in ccRCC cells exhibited the opposite effects from the above results achieved by SOX17 knockdown (Fig. 3f; Fig. S3g, h).
a RNA sequencing was performed in 786-O cells with or without SOX17 knockdown, and a heatmap depicting the significantly differentially expressed genes is presented. b Analysis of pathways in 786-O cells without or with SOX17 knockdown is shown. Real-time PCR assays were used to detect the expression levels of YAP and its target genes in 786-O cells without or with SOX17 knockdown (c) or with SOX17 overexpression (d). Western blot analysis of p-LATS1/2, LATS1/2, p-YAP, and YAP in cytoplasmic (Cyt) and nuclear (Nuc) fractions of 786-O cells without and with SOX17 knockdown (e) or with SOX17 overexpression (f) are presented. β-actin and histone H3 were used to be internal controls for the cytoplasmic and nuclear fractions, respectively. g Luciferase assays were applied to determine the transcriptional activity of YAP in SOX17-knockdown and control 786-O cells without or with the SOX17-binding site on the YAP promoter blocked by the mutant plasmid harboring mutated variants. A schematic representation of the SOX17 consensus and the relative position respect to the TSS are presented. h ChIP-PCR analysis was used to test the binding of SOX17 to YAP promoter in 786-O cells. Real-time PCR assays were applied to examine the expression levels of YAP target genes in 786-O cells and SOX17-knockdown 786-O cells without or with decreasing YAP (i) or with TEAD1 (j) by using siRNAs. CCK-8 assays (k) and flow cytometry assays (l) were performed to test the proliferation (k) and percentage of apoptotic cells (l) of ccRCC cells and SOX17-knockdown ccRCC cells without or with decreasing YAP, respectively. Representative images and statistical analysis of the invasion (m) or migration (n) assays in ccRCC cells and SOX17-knockdown ccRCC cells without or with decreasing YAP are presented (scale bar = 200 µm). o 786-O cells and SOX17-knockdown 786-O cells without or with decreasing YAP were treated with pazopanib (6 μM) or sunitinib (8 μM) and cell proliferation was detected by CCK-8 assays for different times. p, q 786-O cells and SOX17-knockdown 786-O cells without or with decreasing YAP were subcutaneously injected into nude mice (n = 5/group) and the tumor xenografts are presented (p). Representative images of H&E and IHC staining for SOX17, YAP, and Ki-67 in subcutaneous xenografts are presented (p; scale bar = 20 μm). The volumes of tumor xenografts from the three groups were compared at the indicated times (once a week for 6 weeks after inoculation) (q). All p values are defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Then, we examined how SOX17 regulated the transcription of YAP. First, a luciferase promoter activity assay was performed to show that SOX17 knockdown enhanced the transcriptional activity of YAP in ccRCC cells (Fig. 3g; Fig. S3i). Then the online JASPAR software (http://jaspar.genereg.net) was applied to predict the putative SOX17 binding sites of YAP promoter (Fig. S3j). ChIP-qPCR demonstrated that SOX17 bound to the promoter of YAP (Fig. 3h). However, when the SOX17 binding site on the YAP promoter was blocked by the mutant plasmid harboring mutated variants, the upregulated transcriptional activity of YAP induced by SOX17 knockdown was inhibited (Fig. 3g; Fig. S3i). Moreover, SOX17 knockdown or overexpression increased or decreased the expression of TEA domain transcription factor 1 (TEAD1), respectively (Fig. 3c, d; Fig. S3c, d), which is usually recruited by YAP to induce the expression of downstream genes in tumors [22]. In addition, a coimmunoprecipitation (co-IP) assay showed the amounts of the YAP and TEAD1 protein complex were increased in SOX17-knockdown ccRCC cells (Fig. S3k), and knockdown of YAP or TEAD1 abolished the upregulated expression of the target genes of YAP induced by SOX17 knockdown in ccRCC cells (Fig. 3i, j; Fig. S3l–o).
We know that activated LATS1/2 phosphorylates YAP by binding to it, and p-YAP binds to 14-3-3 proteins, which leads to the cytoplasmic sequestration of YAP and the decreased localization of YAP in the nucleus [22]. Then we examined whether SOX17 could directly bind to YAP and LATS1/2 to regulate the LATS1/2-induced phosphorylation of YAP. Accordingly, the co-IP assay demonstrated that there was a direct interaction between endogenous SOX17 and YAP or LATS1/2 (Fig. S3p, q) and the amounts of YAP and pLATS1/2 or p-YAP and 14-3-3 protein complex were decreased in SOX17-knockdown ccRCC cells (Fig. S3r, s). Therefore, SOX17 facilitates the cytoplasmic sequestration of YAP by promoting the LATS1/2-induced phosphorylation of YAP, which inhibits the nuclear distribution of YAP in ccRCC. Moreover, knockdown of YAP or TEAD1 abolished the enhanced proliferation, migration, invasion, targeted drug resistance, and tumorigenicity, and the decreased apoptosis induced by SOX17 knockdown in ccRCC cells (Fig. 3k–q; Fig. S3t–z). Overall, SOX17 plays a tumor suppressive role in ccRCC by inactivating YAP signaling.
SOX17 inhibits the progression of ccRCC by regulating the interaction between ccRCC and TAMs
Recent studies, including ours, have reported that tumor progression depends on not only the intratumoral signaling pathways but also tumor-associated macrophages (TAMs) [17, 18]. As expected, we showed that there was a negative correlation between SOX17 expression and CD163 (a marker for TAMs) [23] in ccRCC specimens (Fig. 4a, b). Then, cohort 1 of ccRCC patients was divided into four groups according to the optimal cut-off values of SOX17 and CD163 (Fig. S1e; Fig. S4a). Among the groups, patients with low SOX17 expression and high CD163 expression exhibited not only unfavorable clinicopathological characteristics but also worse OS and PFS (Fig. 4c; Table S3). Moreover, SOX17 and CD163 were identified as independent risk factors for the prognosis of ccRCC patients (Table S4). Furthermore, a time-dependent c-index analysis demonstrated that the incorporation of both SOX17 and CD163 expression with the clinical indicators TNM stage or SSIGN score exhibited the highest c-index value (Tables S5 and6). The above results were also confirmed in cohort 2 and in the randomized training, validation, and combined cohorts (Fig. 4d, Fig. S4b–d and Tables S5–8).
a Representative images of H&E and IHC staining for SOX17 and CD163 in ccRCC tissues are presented (different scale bars are presented on the images). b The correlation analysis between SOX17 and CD163 in ccRCC is presented (n = 539). c, d Kaplan–Meier analyses for OS and PFS were performed in ccRCC patients from cohort 1 (n = 343; c) or cohort 2 (n = 115; d). Migration assays (e) and real-time PCR assays (f) for U937 cells in the absence or presence of CM from 786-O cells, SOX17-overexpressing 786-O cells without or with concurrent upregulation of YAP are shown. Migration assays (g) and real-time PCR assays (h) for U937 cells in the absence or presence of CM from 786-O cells, SOX17-knockdown 786-O cells without or with concurrently with decreasing YAP are shown. CCK-8 (i), flow cytometry (j), invasion (k), and migration (l) assays were used to detect the proliferation, apoptosis, invasion, and migration of 786-O cells, cocultured 786-O cells in the absence or presence of SOX17 overexpression without or concurrently with YAP upregulation. m 786-O cells, cocultured 786-O cells in the absence or presence of SOX17 overexpression without or concurrently with upregulating YAP were treated with pazopanib (6 μM) and cell proliferation was detected by CCK-8 assays for different times. All p values are defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Then we determined whether intratumoral SOX17 regulated ccRCC-TAM interactions. First, a human leukemic monocyte lymphoma cell line (U937 cells that can be differentiated into macrophages) [24] was exposed to conditioned medium (CM) from ccRCC cells with or without SOX17 overexpression, and a lower number of U937 cells migrating toward ccRCC cells with SOX17 overexpression was observed (Fig. 4e; Fig. S4e, f). In addition, in the presence of SOX17-overexpressing ccRCC CM, U937 cells presented lower expressions of M2-type genes and higher expressions of M1-type genes (Fig. 4f; Fig. S4g). Moreover, the cooverexpression of YAP in ccRCC cells abated the inhibition of migration and M2-type gene expression of U937 cells induced by SOX17 (Fig. 4e, f; Fig. S4f, g). Conversely, CM from ccRCC cells with SOX17 knockdown resulted in increased migration and M2-type genes of U937 cells, while silencing YAP in ccRCC cells alleviated the above promoting effects (Fig. 4g, h; Fig. S4h, i).
In addition, a coculture system was established to determine whether SOX17 regulated the effects of TAMs on the biological behaviors of ccRCC. After the coculture of ccRCC cells and U937 cells, ccRCC cells presented with increased proliferation, decreased apoptosis, and enhanced invasion, migration, and targeted drug resistance, while overexpressed SOX17 in ccRCC cells inhibited the protumoral effects of TAMs (Fig. 4i–m; Fig. S4j–n). However, the simultaneous overexpression of YAP alleviated the inhibitory effects of TAMs on ccRCC cells induced by SOX17 overexpression (Fig. 4i–m; Fig. S4j–n). In contrast, knockdown of SOX17 further enhanced the effect of TAMs on ccRCC cells, while silencing YAP in ccRCC cells alleviated the promoting effects of TAMs on ccRCC cells induced by SOX17 knockdown (Fig. S4o–s). Therefore, decreasing SOX17 in ccRCC promotes the recruitment and activation of TAMs via YAP, which reciprocally facilitates the growth and progression of ccRCC.
CCL5/CCR5 axis is involved in SOX17 regulation of the interaction between ccRCC cells and TAMs
We next investigated the mechanisms underlying the SOX17-mediated regulation of ccRCC-TAM interactions. First, cytokine profiles in the CM from U937 cells, 786-O cells, cocultured U937/786-O cells, and U937/786-O-SOX17-overexpressing cells were examined by a RayBio Human Cytokine Antibody Array (Fig. 5a–c, Fig. S5a, b and Table S9). Compared with the CM from U937 or 786-O cells alone, the CM from cocultured U937/786-O cells almost presented upregulated cytokines in both comparable groups (Fig. 5a, b; Table S9). However, when CM from U937/786-O-SOX17-overexpressing cells and cocultured U937/786-O cells were compared, most of the above cytokines were differentially downregulated (Fig. 5c; Table S9). We then analyzed the comparable groups by using a Venn diagram, which demonstrated that five proteins were significantly expressed (Fig. 5d; Table S9). To select the crucial proteins, metrics and combining both p value and fold change, was used to determine the protein relevance rank. As shown in Fig. S5c and Table S9, C-C motif chemokine 5 (CCL5) was crucial in the SOX17-regulated ccRCC-TAM interaction. In addition, these findings were also validated by ELISA. Although CCL5 in the CM from cocultured U937/ccRCC cells was increased compared with that in the CM from U937 or 786-O cells alone, overexpression or knockdown of SOX17 in ccRCC cells downregulated or further enhanced the CCL5 concentration, respectively, in the CM from U937/786-O cocultured cells (Fig. 5e, f; Fig. S5d–e). Moreover, simultaneously overexpressing or silencing YAP alleviated the effects of TAMs on the CCL5 concentration induced by SOX17 overexpression or knockdown (Fig. 5e, f; Fig. S5d, e). Furthermore, although SOX17 knockdown in ccRCC cells further enhanced the recruitment and activation of TAMs, the addition of a CCL5 neutralizing antibody or an inhibitor of CCR5 (maraviroc, which inhibits the main receptor for CCL5 binding to activate downstream signaling) [25] alleviated the promoting effects of SOX17 knockdown on TAMs (Fig. 5g, h; Fig. S5f, g). In addition, the CCL5 neutralizing antibody or maraviroc abated the increased proliferation, decreased apoptosis, and enhanced invasion, migration, and targeted drug resistance of ccRCC cells after the coculture of U937/ccRCC cells with SOX17 knockdown (Fig. 5i–m; Fig. S5h–l).
Heatmaps of significantly expressed cytokines in CM from different groups were compared as follows: a cocultured U937/786-O with 786-O cells; b cocultured U937/786-O with U937 cells; c cocultured U937/786-O-SOX17-overexpression with U937/786-O. d A Venn diagram of the significantly expressed cytokines of the indicated groups is presented. e ELISAs were used to determine the CCL5 concentration in CM from 786-O cells, U937 cells, cocultured 786-O cells in the absence or presence of SOX17 overexpression without or concurrently with upregulating YAP. f ELISAs were used to determine the CCL5 concentration in CM from 786-O cells, U937 cells, cocultured 786-O cells in the absence or presence of SOX17 knockdown without or concurrently with decreasing YAP or TEAD1. Migration assays (g) and real-time PCR assays (h) for U937 cells alone or cocultured with 786-O cells without or with SOX17 knockdown in the absence or presence of a neutralizing antibody specific for CCL5 or the inhibitor maraviroc are shown. CCK-8 (i), flow cytometry (j), invasion (k), and migration assays (l) were used to detect the proliferation, apoptosis, invasion, and migration of 786-O cells, cocultured 786-O cells without or with SOX17 knockdown in the absence or presence of a neutralizing antibody specific for CCL5 or maraviroc are presented. m 786-O cells, cocultured 786-O cells without or with SOX17 knockdown in the absence or presence of a neutralizing antibody specific for CCL5 or maraviroc were treated with pazopanib (6 μM) and cell proliferation was detected by CCK-8 assays for different times. n Real-time PCR was used to detect the mRNA expression of CCL5 in 786-O cells, cocultured 786-O cells in the absence or presence of SOX17 knockdown without or concurrently with decreasing YAP, TEAD1 or in the presence of verteporfin. Migration assays (o) and real-time PCR assays (p) for U937 cells alone or cocultured with 786-O cells without or with SOX17 knockdown in the absence or presence of decreasing TEAD1 or verteporfin treatment are shown. All p values are defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Next, we examined how SOX17 regulated CCL5 in ccRCC. First, higher CCL5 mRNA expression and transcriptional activity were observed in ccRCC cells after coculture with U937 cells, and SOX17 knockdown further enhanced the expression and transcription of CCL5 (Fig. 5n; Fig. S5m, n). However, when YAP was simultaneously silenced, the SOX17 knockdown-induced upregulation of CCL5 was inhibited (Fig. 5n; Fig. S5m, n), suggesting that SOX17 mediates CCL5 transcription in ccRCC through YAP. Given that YAP is not a transcription factor and often binds with TEAD1 to trigger downstream genes, we first performed a ChIP-PCR assay to demonstrate that TEAD1 could bind to CCL5 promoter (Fig. S5o–p). Blocking the binding sites of the CCL5 promoter by TEAD1 inhibited the SOX17 knockdown-induced enhanced transcriptional activity of CCL5 in cocultured ccRCCs, and this effect was validated by luciferase assays (Fig. S5q). Thus, SOX17 inhibits CCL5 transcription via inhibition of YAP transcriptional function with TEAD1.
Moreover, we further investigated whether knockdown of TEAD1 or interrupting the YAP-TEAD1 combination could alleviate the promoting effects of SOX17 knockdown on TAM-ccRCC cell interactions. First, as shown in Fig. 5n; Fig. S5m–n, S5r, verteporfin (an inhibitor of YAP-TEAD1 binding [26]) treatment both reduces CCL5 expression and the SOX17 knockdown-induced upregulation of CCL5 in ccRCC cells. Although CM from ccRCC cells with SOX17 knockdown resulted in increased migration and M2-type genes in U937 cells, the silencing of TEAD1 or the addition of verteporfin in ccRCC cells abated these promoting effects (Fig. 5o, p; Fig. S5s–t). In addition, after the coculture of ccRCC cells and U937 cells, although knockdown of SOX17 further enhanced the role of TAMs on ccRCC cells, TEAD1 silencing or verteporfin administration to ccRCC cells alleviated the promoting effects (Fig. S5u–z).
CCL5 reciprocally activates CCR5/STAT3 to suppress SOX17 transcription and upregulates YAP to form a positive regulatory loop in the ccRCC-TAM interaction
We further investigated whether CCL5/CCR5 inhibited intratumoral SOX17 and activated YAP signaling in ccRCC. First, recombinant CCL5 promoted the proliferation, invasion, and migration abilities of ccRCC cells in a dose- and time-dependent manner (Fig. 6a–d; Fig. S6a, b). However, when the CCR5 inhibitor maraviroc was added to the CM of recombinant CCL5-treated ccRCC cells, the CCL5-enhanced effects were alleviated (Fig. 6a–d; Fig. S6a, b). Second, decreased SOX17 expression and upregulated expression of YAP and its target genes were observed in ccRCC with recombinant CCL5 treatment compared with naïve ccRCC cells (Fig. 6e; Fig. S6c). In addition, CCL5-treated ccRCC cells exhibited downregulated p-YAP and decreased YAP expression in the cytoplasm, while more YAP appeared in the nucleus (Fig. 6f; Fig. S6d). CCL5 activates downstream STAT3 by binding to CCR5, which was consistent with our results that CCL5 upregulated p-STAT3 in ccRCC (Fig. 6g; Fig. S6e). However, the CCR5 inhibitor maraviroc abated the above effects of CCL5 on ccRCCs (Fig. 6e–g; Fig. S6c–e). Moreover, decreased expression of SOX17, activated YAP signaling, and upregulated p-STAT3 were observed in ccRCC cells after coculture with U937 cells, while the addition of the neutralizing antibody specific for CCL5 or the CCR5 inhibitor maraviroc abated the above effects (Fig. 6h–j; Fig. S6f–h).
ccRCC cells were exposed to a range of concentrations of recombinant human CCL5 protein (15 or 20 ng/ml) for 2 or 3 days in the absence or presence of maraviroc (5 μM), and the proliferation viability (a, b), invasion (c), and migration abilities (d) of ccRCC cells are shown. e Real-time PCR was used to determine the mRNA expression of SOX17, YAP, the target genes of YAP, and TEAD1 in 786-O cells without or with recombinant human CCL5 protein (15 or 20 ng/ml) for 3 days in the absence or presence of maraviroc (5 μM). f Western blot analysis of p-YAP and YAP in cytoplasmic (Cyt) and nuclear (Nuc) fractions of 786-O cells without or with recombinant human CCL5 protein (15 or 20 ng/ml) for 3 days in the absence or presence of maraviroc (5 μM). g Western blot analysis of p-STAT3 and STAT3 in 786-O cells without or with recombinant human CCL5 protein (15 or 20 ng/ml) for 3 days in the absence or presence of maraviroc (5 μM). h Real-time PCR was used to determine the expression of SOX17, YAP, the target genes of YAP, and TEAD1 in 786-O cells alone or cocultured 786-O cells in the absence or presence of a neutralizing antibody specific for CCL5 or maraviroc. i Western blot analysis of p-YAP and YAP in cytoplasmic (Cyt) and nuclear (Nuc) fractions of 786-O cells alone or cocultured 786-O cells in the absence or presence of a neutralizing antibody specific for CCL5 or maraviroc. j Western blot analysis of p-STAT3 and STAT3 in 786-O cells alone or cocultured 786-O cells in the absence or presence of a neutralizing antibody specific for CCL5 or maraviroc. k ChIP-PCR analysis was applied to test the binding of STAT3 to SOX17 promoter in 786-O cells. l Luciferase assays were applied to detect the transcriptional activity of SOX17 in 786-O cells without or with recombinant human CCL5 protein (15 or 20 ng/ml) in the absence or presence of a STAT3-binding site on the SOX17 promoter blocked by the mutant plasmid harboring mutated variants. m Luciferase assays were applied to detect the transcriptional activity of SOX17 in 786-O cells alone or with cocultured 786-O cells in the absence or presence of a STAT3-binding site on the SOX17 promoter blocked by the mutant plasmid harboring mutated variants. All p values are defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Next, we determined how CCL5/CCR5 regulated SOX17 transcription. Given the above results that activated CCL5 triggered STAT3, which serves as a transcription factor, we used the online JASPAR software to predict the putative STAT3 binding sites of SOX17 promoter (Fig. S6i). The ChIP-qPCR assay demonstrated that STAT3 bound to the promoter of SOX17 in 786-O cells (Fig. 6k). Moreover, although CCL5-treated ccRCC cells presented with a lower transcriptional activity of SOX17 than naïve ccRCC cells, blocking the binding site of STAT3 on the SOX17 promoter by the mutant plasmid restored the SOX17 expression inhibited by CCL5 (Fig. 6l; Fig. S6j). In addition, after the coculture of ccRCC cells and U937 cells, the transcription of SOX17 was inhibited compared with that in the control ccRCC cells, but blocking the binding site of STAT3 on the SOX17 promoter alleviated the inhibitory effects (Fig. 6m; Fig. S6k). These results indicate that CCL5/CCR5 inhibits SOX17 transcription via STAT3 and activates YAP signaling in ccRCC.
Blocking the interaction between ccRCC cells and TAMs inhibits the metastasis of target drug-resistant ccRCC
We next examined whether targeting the positive regulatory loop suppressed the metastasis of targeted drug-resistant ccRCC. First, pazopanib- and sunitinib-resistant 786-O cell lines (786-O-PR and 786-O-SR) were both established as previously described in our studies [27, 28]. Then, we showed that after coculture with targeted drug-resistant ccRCC, the migration number and M2-type genes of U937 cells were increased compared with those in control U937 cells without coculture (Fig. 7a, b; Fig. S7a–c). However, the addition of maraviroc or verteporfin inhibited the recruitment and activation of TAMs; moreover, combining the two therapies achieved a better effect (Fig. 7a, b; Fig. S7a–c). Second, although targeted drug-resistant ccRCC cells with U937 cells harbored enhanced invasion and migration abilities, maraviroc or verteporfin treatment, especially the two inhibitors combined, simultaneously abated the promoting effects induced by the ccRCC-TAM interaction (Fig. 7c, d; Fig. S7d–g).
Migration assays (a) and real-time PCR assays (b) were performed in U937 cells alone or cocultured with 786-O-PR cells in the absence or presence of verteporfin, maraviroc or the combination of the two inhibitors. Statistical analysis of the invasion (c) or migration assays (d) in 786-O-PR cells alone or cocultured with U937 cells in the absence or presence of verteporfin, maraviroc or combining the two inhibitors are presented. e, f 786-O-PR cells alone or cocultured 786-O-PR cells (with U937 cells) with stable luciferase expression were injected into the caudal vein of mice, and lung metastases were monitored using an in vivo imaging system. After 3 weeks, the mice injected with cocultured 786-O-PR cells were randomly divided into four groups: treated with normal saline, verteporfin, maraviroc or a combination of the two inhibitors. Photon flux levels were examined in the different groups of mice, and the results are presented as the fold increase in lung metastases (e). Images of luciferase intensity and lung metastases from different groups are presented (f). H&E and IHC staining of SOX17, YAP, CCL5, STAT3, and vimentin in lung metastases (f; scale bar = 20 µm). g 786-O-PR cells or cocultured 786-O-PR cells in the absence or presence of verteporfin, maraviroc or a combination of the two inhibitors were treated with pazopanib (6 μM) and cell proliferation was detected by CCK-8 assays for different times. h, i 786-O-PR cells with stable luciferase expression were was injected under the renal capsule of mice, and tumor growth was monitored using an in vivo imaging system. After 3 weeks, the mice injected with 786-O-PR cells were treated with normal saline or pazopanib (80 mg/kg) in the absence or presence of verteporfin, maraviroc or a combination of the two inhibitors. Photon flux levels were examined in the different groups of mice, and the results are presented as the fold increase in tumor growth (h). Images of luciferase intensity and orthotopic xenografts from different groups are presented. H&E and IHC staining of SOX17, YAP, CCL5, STAT3, and F4/80 in tumor specimens were performed (scale bar = 20 µm) (i). j Schematic diagram of the underlying mechanisms described in our study and the clinical significance of our findings. All p values are defined as *p < 0.05, **p < 0.01, and ***p < 0.001.
Furthermore, 786-O-PR cells labeled with stable luciferase either cocultured with or without U937 cells were injected into the caudal veins of nude mice, and after 3 weeks, the mice were randomly separated into five groups (Fig. 7e, f). Although 786-O-PR cells cocultured with U937 cells caused more serious lung metastasis than 786-O-PR cells alone, maraviroc or verteporfin treatment alleviated lung metastasis induced by the co-culture of 786-O-PR and U937 cells (Fig. 7e, f). Moreover, combining the administration of maraviroc and verteporfin achieved the best inhibitory effect (Fig. 7e, f). In addition, IHC assays demonstrated that the group treated with both maraviroc and verteporfin showed the highest SOX17 expression and lowest expressions of YAP, CCL5, STAT3, and vimentin (Fig. 7f; Fig. S7h–l). Therefore, targeting the regulatory network between ccRCC and TAMs by using maraviroc and verteporfin effectively inhibits the metastasis of targeted drug-resistant ccRCC.
Targeting tumor-TAM interactions by blocking the positive regulatory network SOX17low/YAP/CCL5/CCR3/STAT3 impairs the target drug resistance of ccRCC
Furthermore, we examined whether blocking the positive regulatory loop SOX17low/YAP/CCL5/CCR3/STAT3 could suppress the targeted drug resistance of ccRCC. First, after coculture with U937 cells, 786-O-PR, and 786-O-SR cells exhibited stronger resistance to pazopanib and sunitinib, respectively, than either 786-O-PR or 786-O-SR cells alone (Fig. 7g; Fig. S7m). However, when either maraviroc or verteporfin was added to the cell CM, the drug resistance induced by TAMs was abated (Fig. 7g; Fig. S7m). In particular, the simultaneous addition of maraviroc and verteporfin resulted in the most obvious inhibitory effects (Fig. 7g; Fig. S7m). Thus, blocking the interaction between ccRCC and TAMs reverses the targeted drug resistance of ccRCC.
Furthermore, an orthotopic ccRCC model was established using luciferase-expressing 786-O-PR cells injected into the subcapsular kidneys of NOD/SCID mice. Three weeks postinjection, the mice were randomly separated into five subgroups: (a) naïve 786-O-PR; (b) 786-O-PR with pazopanib treatment; (c) pazopanib-treated 786-O-PR with maraviroc, (d) verteporfin, or (e) the combination of both maraviroc and verteporfin. As shown in Fig. 7h, i, there was no significant difference in tumor growth between the pazopanib-treated 786-O-PR and naïve 786-O-PR groups, indicating the resistance features of 786-O-SR-derived orthotopic ccRCC in vivo. However, compared with the pazopanib-treated and naïve 786-O-PR groups, the pazopanib-treated 786-O-PR group treated with maraviroc or verteporfin presented with reduced tumor growth and decreased tumor volumes (Fig. 7h, i). More importantly, combination therapy by using maraviroc and verteporfin achieved the best therapeutic effects (Fig. 7h, i). In addition, IHC assays demonstrated that the combination therapy group presented with the least TAM infiltration, highest SOX17 expression and lowest YAP, CCL5, and STAT3 expression levels among the groups (Fig. 7i; Fig. S7n–r). Therefore, targeting the tumor-TAM interactions by blocking the positive regulatory loop inhibits the drug resistance of targeted drug-resistant ccRCC.
Discussion
Recently, SOX17 has been reported in various types of cancers and exerts a tumor-suppressive role [29]. Among the previous studies, the mechanisms underlying SOX17 mainly focused on the Wnt/β-catenin pathway [29]. In the present study, we found a novel mechanism by which SOX17 inhibited YAP transcription and facilitated the phosphorylation of YAP by p-LATS1/2 in ccRCC cells. Moreover, SOX17 inhibited the progression of ccRCC by blocking tumor-macrophage interactions via suppressing the SOX17low/YAP/TEAD1/CCL5/CCR5/STAT3 regulatory loop (Fig. 7j).
Many studies, including ours, have demonstrated that both intratumoral signaling pathways and TAMs in the microenvironment persistently trigger protumoral genes and pathways in tumor cells [17, 18, 30]. We also found that intratumoral SOX17 expression was negatively correlated with TAM infiltration in ccRCC specimens. Although other studies reported that ccRCC could educate TAMs and that their interaction promoted the progression of ccRCC [31], further research is needed to clarify the relevant molecular mechanisms. The present study demonstrated that ccRCC cells with decreased intratumoral SOX17 expression activated TAMs by promoting YAP/TEAD1 to trigger CCL5 transcription and expression; then, TAMs reciprocally further inhibited SOX17 and enhanced ccRCC progression via CCL5/CCR5/STAT3. Moreover, although many studies have examined how SOX17 mediates downstream signaling pathways in tumors [6, 7], the upstream mechanism regulating SOX17 expression remains unknown. Our study also showed that CCL5/CCR5-activated STAT3 directly bound to the SOX17 promoter and inhibited SOX17 transcription, which suggests that a CCR5 inhibitor could upregulate SOX17 expression and inhibit ccRCC.
CCL5 is expressed by T lymphocytes, macrophages, platelets, synovial fibroblasts, tubular epithelium, and certain types of tumor cells and is associated with an array of human diseases, including AIDS, autoimmune diseases, and cancer [32]. Though CCL5 can induce immune responses against tumors, CCL5 is also associated with cancer progression and metastasis [33]. Previous studies have shown that CCL5 is associated with carcinogenesis and the prognosis of ccRCC [34], but the biological function of CCL5 in ccRCC remains unknown. In the present study, we demonstrated that CCL5 promoted the malignant features of ccRCC cells via CCR5. In addition, CCL5 reciprocally suppressed SOX17 transcription and upregulated YAP to form a positive regulatory loop in ccRCC.
In conclusion, our results provide a new scientific basis for elucidating the mechanisms underlying the progression and treatment-induced resistance of ccRCC and will help to effectively treat advanced and metastatic ccRCC in the clinic. However, whether SOX17 serves as a tumor-initiating driver and induces an immunosuppressive microenvironment in ccRCC, and why the LATS1/2 transcript is reduced but the total protein is not downregulated upon SOX17 interference needs further study.
Materials and methods
Patients and samples
A total of 539 paired ccRCC tissues and adjacent tissues from patients from four independent clinical centers were used in the present study, and among the patients, two independent cohorts, cohort 1 (n = 343, Changhai Hospital, Shanghai, China) and cohort 2 (n = 115, Fudan University Shanghai Cancer Center, Shanghai, China) of ccRCC patients who were pathologically diagnosed between 2010 and 2014 were recruited for this study to examine the prognostic value of different markers. The clinical characteristics of the two independent cohorts of ccRCC patients are summarized in Table S10. Two pathologists were blinded to the clinicopathological data and scored all samples independently. The 64 paired ccRCC specimens used for real-time PCR and the 30 cases of ccRCC tissues and their corresponding adjacent normal controls used for western blotting were collected from the Gongli Hospital and the Third Affiliated Hospital of Second Military Medical University in China. All patients were pathologically diagnosed with ccRCC between 2017 and 2018. Specimens were collected and stored in liquid nitrogen immediately after surgery. This study followed the recommendations for prognostic studies investigating tumor biomarkers (REMARK) [35]. All experiments were approved by the institutional ethical review board from each hospital, and written informed consent was obtained from all ccRCC patients.
Western blotting and co-IP analysis
Western blotting was performed as described in our previous study [22]. Briefly, whole cell extracts or ccRCC specimens were prepared in RIPA lysis buffer and centrifuged at 12,000 × g for 15 min. Protein concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA). Immunoblotting was performed using specific primary antibodies, overnight in the blocking solution at 4 °C. Then membranes were incubated with the corresponding secondary antibody at room temperature for 1–2 h and detected using the Amersham Imager 600 (General Electric Company, Fairfield, Connecticut, USA). Nuclear and cytoplasmic proteins from ccRCC cells were extracted using an NE-PER Nuclear and Cytoplasmic Extraction Kit (#78899, Thermo Scientific). The primary and secondary antibodies were listed in Table S11.
Coimmunoprecipitation experiments were performed according to previously published protocols [17]. Briefly, the pre-treated 10 μl protein A agarose beads were added to the cell lysate incubated overnight with the antibody and slowly shaken at 4 °C for 2–4 h to attach the antibody to the protein A agarose beads. Then, the agarose bead bound protein was eluted with 1 ml lysis buffer for the next step, and the antibodies used are listed in Table S11.
Real-time polymerase chain reaction (real-time PCR)
Real-time PCR assays were carried out as described in our previous study [36]. Briefly, total RNA from different cell lines and human tissues was extracted using TRIzol Reagent (15596018, Gibco, Grand Island, New York, USA). cDNA was synthesized using the PrimeScript One Step RT Reagent Kit (RR064A, TaKaRa, Kyoto, Japan) following the manufacturer’s instructions. Quantitative RT-PCR was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, California, USA) and SYBR Green Real-Time PCR Master Mix (QPK201, Toyobo, Osaka, Japan). All results were normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The fold change relative to the mean value was determined by the 2−▵▵Ct method. The sequences of primers were presented in Table S11.
Immunohistochemistry (IHC)
The IHC assay was performed as previously reported [17]. Briefly, all tissues were fixed in 4% methanol and embedded in paraffin, and cut into 5-µm-thick sections. Deparaffinization and rehydration were performed according to routine methods, and antigen recovery was performed in heated citrate buffer (pH 6.0) or EDTA buffer (pH 8.0) for 30 min. The tissue microarray slides were then incubated with the UltraSensitive SP (Mouse/Rabbit) IHC Kit (KIT-9710, Maixin Biotechnologies, Fuzhou, China). Specifically, endogenous peroxidases were blocked in each slide by incubating the slides with endogenous peroxidase blockers for 30 min, and non-specific binding sites were blocked with normal animal nonimmune serum for 20 min. The primary antibodies listed in Table S11 were used in the incubation at 4 °C overnight. Then, the slides were incubated with a biotin-labeled secondary antibody and streptomyces antibiotic protein-peroxidase for 30 min separately. Diaminobenzidine (DAB) (DAB-2031, Maixin Biotechnologies) staining was applied. Then, the sections were counterstained with hematoxylin, and quantified by two independent investigators in a double-blinded manner. SOX17, YAP, CCL5, and vimentin staining were scored semiquantitatively as negative (0), weakly positive (1+), moderately positive (2+), or strongly positive (3+) along with the percentage of positive cells. For each observed tissue component (the cytoplasm and nucleus), a summary value referred to as the component H-Score was calculated by multiplying the intensity score (ranging from 0 to 3) and the percentage of positive cells (ranging from 0 to 300). The total H-Score for a tissue section was derived as the sum of the component H-Scores weighted by the fraction of each component observed in the tissue section, as described in our previous study [37]. Briefly, the whole slide was first observed under a low-power microscope (×100 magnification). Then, three random representative fields of view were selected under a high-power microscope (×200 magnification) and scored according to the previous grading method. Subsequently, the mean value was calculated. IHC staining for STAT3 and Ki-67 were evaluated by the percentage of positive cells. To determine the number of stained cells in each observed tissue component, three respective areas of the tumor core were evaluated at a ×400 magnification, and the mean value was adopted. For TAMs, the whole slide was also first observed at a 100 × magnification. Then, three randomly representative areas of the tumor were evaluated at a ×200 magnification to score the density of stained stromal immune cells. Finally, the mean value was calculated. The total cell count was defined as the number of nucleated stained cells per field and is presented as the density (cells/mm2) [23].
Cell culture
The cell lines used in the present study were obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) in 2018. HK-2 cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (11995-065, Gibco). ACHN cells were cultured in Minimum Essential Medium (11095-080, Gibco), and 786-O, 769-P and OS-RC-2 cells were maintained in RPMI-1640 medium (22400-089, Gibco). The culture media of all cell lines were supplemented with fetal bovine serum (FBS, 10%, Gibco) and 1% penicillin/streptomycin (Gibco), and U937 cells were maintained in RPMI-1640 medium (22400-089, Gibco) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and 0.05 mM β-mercaptoethanol (07604, Sigma, St. Louis, Missouri, USA). Sunitinib- and pazopanib-resistant 786-O cell lines were maintained in RPMI-1640 medium supplemented with 10% (v/v) FBS and 10 µM sunitinib or 8 µM pazopanib. All cell lines were cultured at 37 °C in 5% CO2.
All cell lines in the present study were authenticated by short tandem repeat (STR) profiling and examined for mycoplasma contamination using a Mycoplasma Detection Kit (Selleck Chemicals, Houston, Texas, USA), and the most recent tests were conducted in November 2019. All cell lines used in the study were cultured within 40 passages.
Coculture assays
The coculture assays were performed as previously reported [17]. Briefly, U937 cells were pelleted via centrifugation at 300 × g. Thereafter, the pelleted cells were resuspended in the medium as described above and laid on top of the monolayer of ccRCC cells. After 5 days of coculture at 37 °C and 5% CO2, both the supernatant and cells were collected for subsequent analysis.
Cell proliferation and assessment of apoptosis and cell cycle
The proliferation of ccRCC cells under the indicated conditions was detected using a CCK8 kit (CK-04, Dojindo, Kumamoto, Kyushu, Japan) according to the manufacturer’s instructions, as described in our previous study [38]. Prior to the assay, the medium was replaced with fresh medium, 10% v/v CCK-8 was added to each well, and the samples were incubated at 37 °C for 2 h. The OD values were measured at an absorbance of 450 nm using a microplate reader (EXL800, BioTek Instruments, Winooski, VT, USA). The proliferation rates are presented as proportions of the control value, which was obtained from the control group.
Apoptotic cells were evaluated by annexin-V and propidium iodide (PI) staining (70-APCC101-100, MultiSciences, Hangzhou, China) according to the manufacturer’s instructions, and then analyzed by flow cytometry with a Cyan ADP Sorter (Beckman, CA, USA).
Cell cycle assays were performed according to the manufacturer’s instructions. Cells were stained with propidium iodide (PI) (70-APCC101-100, MultiSciences, Hangzhou, China), and incubated in the dark at room temperature for 30 min. Then PI fluorescence signals were assessed by flow cytometry with a Cyan ADP Sorter (Beckman, CA, USA). Cell cycle distribution was analyzed using Flow Jo software, and gated cells in G1, S or G2/M-phase were counted.
Invasion and migration assays
Invasion and migration assays were conducted in transwell chambers (Millipore, Billerica, MA, USA) with or without Matrigel (BD Biosciences, NJ, USA) according to the manufacturer’s instructions, as described in our previous study [17]. A total of 1 × 104 786-O or 769-P cells or 1 × 105 U937 cells were seeded in RPMI-1640 medium without fetal bovine serum (FBS) into the upper chamber of each uncoated transwell, RPMI-1640 medium with 20% FBS and conditioned medium (CM) was placed in the lower chamber. 36 h or 48 h after seeding, the noninvasive cells in the upper chamber were removed with a cotton swab, and the cells on the lower surface of the membrane were fixed with 4% Paraformaldehyde Fix Solution (E672002, Sangon Biotech, Shanghai, China), stained with crystal violet (E607309, Sangon Biotech), and photographed at ×200 magnification. The data are presented as the means ± SD from three independent experiments.
RNA-Seq and analysis
RNA was isolated from 786-O cells without and with SOX17-knockdown with TRIZOL reagent (Invitrogen, Grand Island, NY). The total RNA was purified by the Qiagen RNeasy mini kit (Qiagen, velencia, CA) and then the purified RNA was checked to determine the quantity. Base calling was performed on an Illumina BaseSpace platform, using the FASTQ Generation Application. Adaptor sequences were removed and low-quality bases were trimmed by using trimmomatic (v0.35). Reads shorter than 15 base pairs were excluded from subsequent analysis. Read sequences were then aligned to the human reference genome GRCh37 using tophat2 (v2.1.1) with GENCODE version 24 gene models. Gene expression levels were inferred from BAM files using htseq-count from HTSeq (v1.3.0). Differential expression analysis between different conditions was performed using DESeq2 (v1.12.4). Potential Gene Ontology enrichment and involvement on biological pathways of genes were examined using the R package goProfiles (v3.6).
Antibody-microarray experiment and Enzyme-linked immunosorbent assay (ELISA)
Cytokine profiles were measured by Quantibody Human Inflammatory Array 3 (QAH-INF-3, RayBiotech, Norcross, GA, USA) by using Sandwich-based Design Principle according to the manufacturer’s instructions. Multiple specific capture antibodies of the inflammatory factors were bound to glass surfaces by the manufacturer. After incubation with the plasma samples, the target inflammatory factors were trapped on the solid surface. Then biotin-labeled detection antibodies specific for different inflammatory factors were added to recognize different isotope of the target factors. The inflammatory factors were selected based on the following criteria: (1) With highly significant differences in expression (p value < 0.005) between exposed and control group. (2) A synthetic score, cumulative product of log10 (p value), was used to rank the intersected important factors among the comparisons, and then top N factors were determined as most important factors.
The CCL5 concentration in cell culture medium or blood serum was measured using an ELISA Kit for CCL5 (RK00077, ABclonal, China) according to the manufacturer’s instructions.
Immunofluorescence analysis
Cells were fixed in 4% Paraformaldehyde Fix Solution (E672002, Sangon Biotech, Shanghai, China) for 15 min at room temperature. Then, the cells were permeabilized with 4‰ Triton X-100 (A110694, Sangon Biotech) for 10 min. After a 30 min incubation with tris-buffered saline solution containing 5% bovine serum albumin (BSA), samples were immunostained with a rabbit anti-YAP1 (ab52771) from Abcam (Cambridge, MA, USA) at 4 °C overnight. Subsequently, samples were incubated with an Alexa Fluor 488-conjugated goat anti-rabbit antibody (#4412 S, Cell Signaling Technology, Boston, Massachusetts, USA). Nuclei were stained with DAPI (E607303, Sangon Biotech). Fluorescence images were observed and collected under a Leica DM5000B fluorescence microscope (Leica, Wetzlar, Hesse, Germany).
Gene knockdown and overexpression
The method of this section was done as we previously reported [18]. After 786-O and 769-P cell lines were cultured in 6-well plates, inoculated at a density of 5 × 104 cells/ml, the cells were transduced with the SOX17-overexpressing lentiviral vectors, shRNA-expressing lentivirus (sh-SOX17) or control lentivirus. 72 h after transduction, the cells were observed and photographed under a microscope. Stable 786-O and 769-P cell lines in which SOX17 was knocked down or overexpression were generated using lentiviral constructs. The sequences of shRNAs and SOX17-overexpressing lentiviral vectors are presented in Table S11. siRNA and plasmid transfection was carried out using Lipofectamine 3000 reagents (L3000015, Invitrogen) according to the manufacturer’s protocols, the sequences of si-YAP, si-TEAD1, and YAP plasmid are shown in Table S11.
Chromatin immunoprecipitation (ChIP) analysis and Luciferase reporter assay
ChIP assays were performed according to our previously published study [18]. Antibodies were listed in Table S11. Primers complementary to the promoter region were used for the detection of YAP, CCL5 and SOX17 genomic DNA, and primers specific to the human GAPDH promoter were used as the control (kit supplied). The enrichment of the targets was calculated as follows: fold enrichment = 2^(Ct [PDGFB-ChIP] - Ct [IgG]). The primer sequences are presented in Table S11.
The sequence of SOX17 binding sites for YAP promoter is 5′-TCCATTGAC-3′, −1736 to −1728 relative to the transcription start site. The sequence of TEAD1 binding sites of the CCL5 promoter is 5′-CTCATTCCAA-3′, −584 to −575 relative to the transcription start site. The sequence of STAT3 binding sites of the SOX17 promoter is 5′-ATTCTAGGAAA-3′, −2038 to −2028 relative to the transcription start site. Mutant constructs related to three promoters were generated using a QuikChange XL site-directed mutagenesis kit (Stratagene), which inducted deletion mutations in promoters. The deleted sequence of YAP promoter, CCL5 promoter, and SOX17 promoter are 5′-CATTGACTC-3′ (−1734 to −1726), 5′-GACTCATTCCAA-3′ (−586 to 575) and 5′-ATTCTAGGAAA-3′ (−2038 to −2028), respectively. The promoter and its mutant sequence were separately cloned into the KpnI and XhoI sites of the pGL3-basic reporter plasmid (Promega, USA). ccRCC cells were cotransfected with 10 ng of the pTK-RL reporter control plasmid and 200 ng of pGL3-basic-YAP (or CCL5, or SOX17)-WT or pGL3-basic-YAP (or CCL5, or SOX17)-Mut using the Lipofectamine 3000 reagent (L3000015, Invitrogen, Waltham, MA, USA) according to the manufacturer’s recommended protocols. The sequences of above plasmids are presented in Table S11. Cells were collected 48 h after transfection, and the YAP (or CCL5, or SOX17) transcriptional activity was evaluated by measuring the luminescence using a Dual-Luciferase Assay Kit (E1910, Promega, Fitchburg, Wisconsin, USA). The fold-induction levels were derived relative to the normalized reporter activity.
Animal experiments
All experimental animal procedures were approved by the Animal Care and Use Committee of Gongli Hospital (Shanghai, China). For the subcutaneous tumor formation assay, 786-O cells (5 × 106 cells in 100 μl of PBS) subjected to different treatments were injected subcutaneously into the nude mice. The mice were euthanized 6 weeks after inoculation, and the tumors were removed, fixed in 10% buffered formalin solution and then used for further experiments. In addition, control 786-O cells, SOX17-knockdown, or SOX17-knockdown with siYAP1# or siTEAD11# (5 × 106 cells in 100 μl of PBS) were injected subcutaneously into the right flank of 6-week-old male nude mice. Two weeks after inoculation, the SOX17-knockdown group was treated with si-NC mixed with in vivo jetPEI reagent (10 μg siRNA/20 μl jetPEI) (Polyplus-transfection, Strasbourg, France), while the SOX17-knockdown with YAP or TEAD1 interference group was treated with siYAP1# or siTEAD11# mixed with in vivo jetPEI reagent. These reagents were injected into the established subcutaneous tumors every 3 days. In addition, the mice were euthanized 6 weeks after inoculation, after which the tumors were removed for IHC assays to validate that the expression of YAP or TEAD1 was low/- in the SOX17-knockdown with YAP/TEAD1 interference group.
For the caudal vein injection of 6-week-old male NOD-SCID mice, 200 μl of PBS containing 1 × 107 786-O-PR-luc cells from the indicated groups was injected into the caudal veins of the mice using a 24-gauge needle. Three weeks after injection, the mice were divided into five groups. In the first group, the mice injected with 786-O-PR cells were treated with normal saline. And mice in the other four groups were injected with 786-O-PR cells that were cocultured with U937 cells in the absence or presence of a CCR5 inhibitor (maraviroc, USA) (10 mg/kg, twice weekly), or a YAP inhibitor (verteporfin, USA) (100 mg/kg every 2 days), or combining use of these two inhibitors. The mice were sacrificed 8 weeks after drug injection, and the lung tissues were removed and fixed in 10% buffered formalin solution.
786-O-PR cells were transfected with the luciferase reporter gene and then used in subcapsular renal tumor formation or lung metastasis assays. After the intraperitoneal injection of D-luciferin (150 mg/kg) (Gold Biotech, USA) in 100 μl of PBS, tumor growth was monitored weekly by live-animal bioluminescence optical imaging using an IVIS Lumina II imaging system (PerkinElmer, Hopkinton, MA, USA). Briefly, for renal subcapsular tumor cell implantation, 6-week-old male NOD-SCID mice were anesthetized and placed in the left lateral decubitus position. A vertical incision was made in the right flank through the skin and peritoneum to expose the lateral aspect of the kidney. The kidney was lifted gently and stabilized, and 50 μl of a Matrigel/medium (1:1) suspension containing 1 × 107 786-O-PR-luc cells from the indicated groups was inoculated under the renal capsule using a 24-gauge needle inserted into the lower pole of the kidney. Three weeks after tumor implantation, the mice were divided into five groups. In two of the groups, the mice injected with 786-O-PR cells maraviroc were treated with normal saline or pazopanib (80 mg/kg). The other three groups were treated with pazopanib (80 mg/kg) in the presence of maraviroc (10 mg/kg, twice weekly), or verteporfin (100 mg/kg every 2 days), or combining use of these two inhibitors. The mice were sacrificed 8 weeks after drug injection, and the normal and injected kidneys were removed and fixed in 10% buffered formalin solution.
Statistical analysis
Numerical data are expressed as the means ± standard deviation (SD). Statistical differences between variables were analyzed by a two-tailed Wilcoxon test. Survival curves were plotted using the Kaplan–Meier method and compared via log-rank analysis. Variables with p values < 0.1 in the univariate analysis were included in the multivariate Cox proportional hazards analysis. Differences were considered significant at p < 0.05. Time-dependent receiver operating characteristic (ROC) analysis was used to determine the cut-off value of SOX17 and CD163 AUC was computed with the “time ROC” package. The prognostic accuracy of the SOX17-CD163 classifier and other prognostic indicators was assessed by Harrell’s concordance index (c-index). All experiments were performed independently at least three times. All the analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc.), SPSS 17.0 (IBM Corporation, Armonk, New York, USA) software, and R software (version 3.5.3).
References
Wei JH, Feng ZH, Cao Y, Zhao HW, Chen ZH, Liao B, et al. Predictive value of single-nucleotide polymorphism signature for recurrence in localised renal cell carcinoma: a retrospective analysis and multicentre validation study. Lancet Oncol. 2019;20:591–600.
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Prim. 2017;3:17009.
Allison TF, Smith AJH, Anastassiadis K, Sloane-Stanley J, Biga V, Stavish D, et al. Identification and single-cell functional characterization of an endodermally biased pluripotent substate in human embryonic stem cells. Stem Cell Rep. 2018;10:1895–907.
Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416–20.
Zhang L, Jambusaria A, Hong Z, Marsboom G, Toth PT, Herbert BS, et al. SOX17 regulates conversion of human fibroblasts into endothelial cells and erythroblasts by dedifferentiation into CD34+ progenitor cells. Circulation. 2017;135:2505–23.
Li L, Yang WT, Zheng PS, Liu XF. SOX17 restrains proliferation and tumor formation by down-regulating activity of the Wnt/β-catenin signaling pathway via trans-suppressing β-catenin in cervical cancer. Cell Death Dis. 2018;9:741.
Merino-Azpitarte M, Lozano E, Perugorria MJ, Esparza-Baquer A, Erice O, Santos-Laso Á, et al. SOX17 regulates cholangiocyte differentiation and acts as a tumor suppressor in cholangiocarcinoma. J Hepatol. 2017;67:72–83.
Kuo IY, Wu CC, Chang JM, Huang YL, Lin CH, Yan JJ, et al. Low SOX17 expression is a prognostic factor and drives transcriptional dysregulation and esophageal cancer progression. Int J Cancer. 2014;135:563–73.
Chen HL, Chew LJ, Packer RJ, Gallo V. Modulation of the Wnt/beta-catenin pathway in human oligodendroglioma cells by Sox17 regulates proliferation and differentiation. Cancer Lett. 2013;335:361–71.
Zhang W, Glöckner SC, Guo M, Machida EO, Wang DH, Easwaran H, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2018;68:2764–72.
Ye YW, Wu JH, Wang CM, Zhou Y, Du CY, Zheng BQ, et al. Sox17 regulates proliferation and cell cycle during gastric cancer progression. Cancer Lett. 2011;307:124–31.
Pascual-García M, Bonfill-Teixidor E, Planas-Rigol E, Rubio-Perez C, Iurlaro R, Arias A, et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8+ T cell tumor-infiltration impairing anti-PD1 therapy. Nat Commun. 2019;10:2416.
Seo N, Shirakura Y, Tahara Y, Momose F, Harada N, Ikeda H, et al. Activated CD8+ T cell extracellular vesicles prevent tumour progression by targeting of lesional mesenchymal cells. Nat Commun. 2018;9:435.
Eruslanov E, Stoffs T, Kim WJ, Daurkin I, Gilbert SM, Su LM, et al. Expansion of CCR8(+) inflammatory myeloid cells in cancer patients with urothelial and renal carcinomas. Clin Cancer Res. 2013;19:1670–80.
Xu L, Zhu Y, Chen L, An H, Zhang W, Wang G, et al. Prognostic value of diametrically polarized tumor-associated macrophages in renal cell carcinoma. Ann Surg Oncol. 2014;21:3142–50.
Di Mitri D, Mirenda M, Vasilevska J, Calcinotto A, Delaleu N, Revandkar A, et al. Re-education of tumor-associated macrophages by CXCR2 blockade drives senescence and tumor inhibition in advanced prostate cancer. Cell Rep. 2019;28:2156–68.
Huang H, Wang C, Liu F, Li HZ, Peng G, Gao X, et al. Reciprocal network between cancer stem-like cells and macrophages facilitates the progression and androgen deprivation therapy resistance of prostate cancer. Clin Cancer Res. 2018;24:4612–26.
Wang C, Peng G, Huang H, Liu F, Kong DP, Dong KQ, et al. Blocking the feedback loop between neuroendocrine differentiation and macrophages improves the therapeutic effects of enzalutamide (MDV3100) on prostate cancer. Clin Cancer Res. 2018;24:708–23.
Roumenina LT, Daugan MV, Noé R, Petitprez F, Vano YA, Sanchez-Salas R, et al. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol Res. 2019;7:1091–105.
Chittezhath M, Dhillon MK, Lim JY, Laoui D, Shalova IN, Teo YL, et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 2014;41:815–29.
Zanconato F, Cordenonsi M, Piccolo S. YAP and TAZ: a signalling hub of the tumour microenvironment. Nat Rev Cancer. 2019;19:454–64.
Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17.
Fu H, Zhu Y, Wang Y, Liu Z, Zhang J, Xie H, et al. Identification and validation of stromal immunotype predict survival and benefit from adjuvant chemotherapy in patients with muscle-invasive bladder cancer. Clin Cancer Res. 2018;24:3069–78.
Wang HC, Chen CW, Yang CL, Tsai IM, Hou YC, Chen CJ, et al. Tumor-associated macrophages promote epigenetic silencing of gelsolin through DNA methyltransferase 1 in gastric cancer cells. Cancer Immunol Res. 2017;5:885–97.
Nie Y, Huang H, Guo M, Chen J, Wu W, Li W, et al. Breast phyllodes tumors recruit and repolarize tumor-associated macrophages via secreting CCL5 to promote malignant progression, which can be inhibited by CCR5 inhibition therapy. Clin Cancer Res. 2019;25:3873–86.
Garcia-Rendueles ME, Ricarte-Filho JC, Untch BR, Landa I, Knauf JA, Voza F, et al. NF2 loss promotes oncogenic RAS-induced thyroid cancers via YAP-dependent transactivation of RAS proteins and sensitizes them to MEK inhibition. Cancer Discov. 2015;5:1178–93.
Wang C, Wang Y, Hong T, Cheng B, Gan S, Chen L, et al. Blocking the autocrine regulatory loop of Gankyrin/STAT3/CCL24/CCR3 impairs the progression and pazopanib resistance of clear cell renal cell carcinoma. Cell Death Discov. 2020;11:117.
Wang C, Li Y, Chu CM, Zhang XM, Ma J, Huang H, et al. Gankyrin is a novel biomarker for disease progression and prognosis of patients with renal cell carcinoma. EbioMed. 2019;39:255–64.
Tan DS, Holzner M, Weng M, Srivastava Y, Jauch R. SOX17 in cellular reprogramming and cancer. Semin Cancer Biol. 2019;S1044-579X:30030–6.
Chen C, He W, Huang J, Wang B, Li H, Cai Q, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9:3826.
Fu Q, Xu L, Wang Y, Jiang Q, Liu Z, Zhang J, et al. Tumor-associated macrophage-derived interleukin-23 interlinks kidney cancer glutamine addiction with immune evasion. Eur Urol. 2019;75:752–63.
Krensky AM, Ahn YT. Mechanisms of disease: regulation of RANTES (CCL5) in renal disease. Nat Clin Pr Nephrol. 2007;3:164–70.
Walens A, DiMarco AV, Lupo R, Kroger BR, Damrauer JS, Alvarez JV. CCL5 promotes breast cancer recurrence through macrophage recruitment in residual tumors. Elife 2019;8:e43653.
Song E, Song W, Ren M, Xing L, Ni W, Li Y, et al. Identification of potential crucial genes associated with carcinogenesis of clear cell renal cell carcinoma. J Cell Biochem. 2018;119:5163–174.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor MARKer prognostic studies (REMARK). Nat Clin Pr Urol. 2005;2:416–22.
Zhao Y, Lu Q, Li C, Wang X, Jiang L, Huang L, et al. PRMT1 regulates the tumour-initiating properties of esophageal squamous cell carcinoma through histone H4 arginine methylation coupled with transcriptional activation. Cell Death Dis. 2019;10:359.
Wang M, Li X, Zhang J, Yang Q, Chen W, Jin W, et al. AHNAK2 is a novel prognostic marker and oncogenic protein for clear cell renal cell carcinoma. Theranostics 2017;7:1100–13.
Wang KJ, Wang C, Dai LH, Yang J, Huang H, Ma XJ, et al. Targeting an autocrine regulatory loop in cancer stem-like cells impairs the progression and chemotherapy resistance of bladder cancer. Clin Cancer Res. 2019;25:1070–86.
Acknowledgements
This work was supported by The Top-level Clinical Discipline Project of Shanghai Pudong (PWYgf2018-03), National Natural Science Foundation of China (No. 81773154, 81772747, 81974391), Shanghai Natural Science Foundation (20ZR1449600), Pudong New Area Science and technology development fund special fund for people’s livelihood Research (medical and health) (PKJ2019-Y19), the Program of Shanghai Academic/Technology Research Leader (No. 19XD1405100), Shanghai key medical specialty project (ZK2019A09), the Shanghai “Rising Stars of Medical Talent” Youth Development Program: Outstanding Youth Medical Talents (XC), Natural Science Foundation of Shanghai (19ZR1447200), Meng Chao Talent Training Program-Cultivation of Leading Talents Reserve (XC), and the Shanghai Medical Guidance (Chinese and Western Medicine) Science and Technology Support Project (No. 17411960200). We thank Dr. Aiping Zhang (Gongli Hospital, Shanghai, China), Yongwei Yu (Changhai hospital, Shanghai, China), and Qifeng Wang (Fudan University Shanghai Cancer Center, Shanghai, China) for their assistance in providing the clinical samples of ccRCC patients. We also thank Shanghai BioGenius biotech. Co., ltd (China) for his BioGenius Cloud Computing Service and bioinformatics analysis.
Author information
Authors and Affiliations
Contributions
CW and XC designed the study; CW wrote the manuscript; CW, YW, TH, JY, CC, LZ, and JZ analyzed the data, performed the experiments, and statistical analysis; XC supervised the study and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M. Piacentini
Rights and permissions
About this article
Cite this article
Wang, C., Wang, Y., Hong, T. et al. Targeting a positive regulatory loop in the tumor-macrophage interaction impairs the progression of clear cell renal cell carcinoma. Cell Death Differ 28, 932–951 (2021). https://doi.org/10.1038/s41418-020-00626-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-020-00626-6
This article is cited by
-
Glycolysis-related biomarker TCIRG1 participates in regulation of renal cell carcinoma progression and tumor immune microenvironment by affecting aerobic glycolysis and AKT/mTOR signaling pathway
Cancer Cell International (2023)
-
Single-cell RNA-seq integrated with multi-omics reveals SERPINE2 as a target for metastasis in advanced renal cell carcinoma
Cell Death & Disease (2023)
-
FCER1G positively relates to macrophage infiltration in clear cell renal cell carcinoma and contributes to unfavorable prognosis by regulating tumor immunity
BMC Cancer (2022)
-
Tumor-associated macrophage-derived exosomes transmitting miR-193a-5p promote the progression of renal cell carcinoma via TIMP2-dependent vasculogenic mimicry
Cell Death & Disease (2022)
-
BAP1 in cancer: epigenetic stability and genome integrity
Discover Oncology (2022)