Abstract
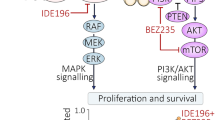
We previously demonstrated that pan-HDAC inhibitors could limit escape from MEK inhibitor (MEKi) therapy in uveal melanoma (UM) through suppression of AKT and YAP/TAZ signaling. Here, we focused on the role of specific HDACs in therapy adaptation. Class 2 UM displayed higher expression of HDACs 1, 2, and 3 than Class 1, whereas HDACs 6, 8, and 11 were uniformly expressed. Treatment of UM cells with MEKi led to modulation of multiple HDACs, with the strongest increases observed in HDAC11. RNA-seq analysis showed MEKi to decrease the expression of multiple HDAC11 target genes. Silencing of HDAC11 significantly reduced protein deacetylation, enhanced the apoptotic response to MEKi and reduced growth in long-term colony formation assays across multiple UM cell lines. Knockdown of HDAC11 led to decreased expression of TAZ in some UM cell lines, accompanied by decreased YAP/TAZ transcriptional activity and reduced expression of multiple YAP/TAZ target genes. Further studies showed this decrease in TAZ expression to be associated with increased LKB1 activation and modulation of glycolysis. In an in vivo model of uveal melanoma, silencing of HDAC11 limited the escape to MEKi therapy, an effect associated with reduced levels of Ki67 staining and increased cleaved caspase-3. We have demonstrated a novel role for adaptive HDAC11 activity in UM cells, that in some cases modulates YAP/TAZ signaling leading to MEKi escape.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Landreville S, Agapova OA, Harbour JW. Emerging insights into the molecular pathogenesis of uveal melanoma. Future Oncol. 2008;4:629–36.
Schank TE, Hassel JC. Immunotherapies for the Treatment of Uveal Melanoma-History and Future. Cancers (Basel). 2019;11:1048.
Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–8.
Field MG, Harbour JW. Recent developments in prognostic and predictive testing in uveal melanoma. Curr Opin Ophthalmol. 2014;25:234–9.
Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602.
Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N. Engl J Med. 2010;363:2191–9.
Vader MJC, Madigan MC, Versluis M, Suleiman HM, Gezgin G, Gruis NA, et al. GNAQ and GNA11 mutations and downstream YAP activation in choroidal nevi. Br J Cancer. 2017;117:884–7.
Musi E, Ambrosini G, de Stanchina E, Schwartz GK. The phosphoinositide 3-kinase alpha selective inhibitor BYL719 enhances the effect of the protein kinase C inhibitor AEB071 in GNAQ/GNA11-mutant uveal melanoma cells. Mol Cancer Ther. 2014;13:1044–53.
Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 2014;25:822–30.
Yoo JH, Shi DS, Grossmann AH, Sorensen LK, Tong Z, Mleynek TM, et al. ARF6 Is an Actionable Node that Orchestrates Oncogenic GNAQ Signaling in Uveal Melanoma. Cancer Cell. 2016;29:889–904.
Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell. 2013;49:94–108.
Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–45.
Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. Jama. 2014;311:2397–405.
Carvajal RD, Piperno-Neumann S, Kapiteijn E, Chapman PB, Frank S, Joshua AM, et al. Selumetinib in Combination With Dacarbazine in Patients With Metastatic Uveal Melanoma: A Phase III, Multicenter, Randomized Trial (SUMIT). J Clin Oncol. 2018;36:1232–9.
Faiao-Flores F, Emmons MF, Durante MA, Kinose F, Saha B, Fang B, et al. HDAC Inhibition Enhances the In Vivo Efficacy of MEK Inhibitor Therapy in Uveal Melanoma. Clin Cancer Res. 2019;25:5686–701.
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20.
Emmons MF, Faiao-Flores F, Sharma R, Thapa R, Messina JL, Becker JC, et al. HDAC8 Regulates a Stress Response Pathway in Melanoma to Mediate Escape from BRAF Inhibitor Therapy. Cancer Res. 2019;79:2947–61.
Landreville S, Agapova OA, Matatall KA, Kneass ZT, Onken MD, Lee RS, et al. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res. 2012;18:408–16.
Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl J Med. 2012;367:107–14.
Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun. 2020;11:496.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–902. e21
Gundersen GW, Jones MR, Rouillard AD, Kou Y, Monteiro CD, Feldmann AS, et al. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics. 2015;31:3060–2.
Faiao-Flores F, Alves-Fernandes D, Pennacchi PC, Sandri S. Vicente ALSA, Scapulatempo-Neto C, et al. Targeting the hedgehog transcription factors GLI1 and GLI2 restores sensitivity to vemurafenib-resistant human melanoma cells. Oncogene. 2017;36:1849.
Paraiso KH, Haarberg HE, Wood E, Rebecca VW, Chen YA, Xiang Y, et al. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin Cancer Res. 2012;18:2502–14.
Faiao-Flores F, Suarez JA, Soto-Cerrato V, Espona-Fiedler M, Perez-Tomas R, Maria DA. Bcl-2 family proteins and cytoskeleton changes involved in DM-1 cytotoxic effect on melanoma cells. Tumour Biol. 2013;34:1235–43.
Bi L, Ren Y, Feng M, Meng P, Wang Q, Chen W, et al. HDAC11 Regulates Glycolysis through the LKB1/AMPK Signaling Pathway to Maintain Hepatocellular Carcinoma Stemness. Cancer Res. 2021;81:2015–28.
Smalley KSM. A pivotal role for ERK in the oncogenic behaviour of malignant melanoma? Int J Cancer. 2003;104:527–32.
Maertens O, Kuzmickas R, Manchester HE, Emerson CE, Gavin AG, Guild CJ, et al. MAPK Pathway Suppression Unmasks Latent DNA Repair Defects and Confers a Chemical Synthetic Vulnerability in BRAF-, NRAS-, and NF1-Mutant Melanomas. Cancer Discov. 2019;9:526–45.
Levinzon L, Madigan M, Nguyen V, Hasic E, Conway M, Cherepanoff S. Tumour Expression of Histone Deacetylases in Uveal Melanoma. Ocul Oncol Pathol. 2019;5:153–61.
Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–55.
Liu SS, Wu F, Jin YM, Chang WQ, Xu TM. HDAC11: a rising star in epigenetics. Biomed Pharmacother. 2020;131:110607.
Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100.
Thole TM, Lodrini M, Fabian J, Wuenschel J, Pfeil S, Hielscher T, et al. Neuroblastoma cells depend on HDAC11 for mitotic cell cycle progression and survival. Cell Death Dis. 2017;8:e2635.
Gong D, Zeng Z, Yi F, Wu J. Inhibition of histone deacetylase 11 promotes human liver cancer cell apoptosis. Am J Transl Res. 2019;11:983–90.
Sun L, Marin de Evsikova C, Bian K, Achille A, Telles E, Pei H, et al. Programming and Regulation of Metabolic Homeostasis by HDAC11. EBioMedicine. 2018;33:157–68.
DeRan M, Yang J, Shen CH, Peters EC, Fitamant J, Chan P, et al. Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 2014;9:495–503.
Wang W, Xiao ZD, Li X, Aziz KE, Gan B, Johnson RL, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17:490–9.
Santinon G, Pocaterra A, Dupont S. Control of YAP/TAZ Activity by Metabolic and Nutrient-Sensing Pathways. Trends Cell Biol. 2016;26:289–99.
Chua V, Han A, Bechtel N, Purwin TJ, Hunter E, Liao C, et al. The AMP-dependent kinase pathway is upregulated in BAP1 mutant uveal melanoma. Pigment Cell Melanoma Res. 2021;35:78–87.
Acknowledgements
This work was supported by NCI/NIH R01 CA256193 to KSMS, JDL, and JWH.
Author information
Authors and Affiliations
Contributions
SNS, FF-F, MFE, BS, CW performed the experiments. SNS, MFE, SC, JDL, IS, MAD, JWH, and KSMS analyzed the data. SNS, MFE, IS, and KSMS wrote the manuscript. MAD, JWH, JDL, and KSMS edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sriramareddy, S.N., Faião-Flores, F., Emmons, M.F. et al. HDAC11 activity contributes to MEK inhibitor escape in uveal melanoma. Cancer Gene Ther 29, 1840–1846 (2022). https://doi.org/10.1038/s41417-022-00452-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-022-00452-7