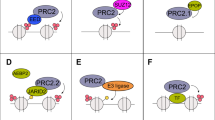
Abstract
Background
Histone modifications alter transcriptional gene function and participate in cancer progression. Enhancer-of-Zeste-Homologue-2 (EZH2) and Nuclear-Receptor-Binding-SET-domain2 (NSD2) methylate H3K27 and H3K36, respectively, to regulate transcription. Given the therapeutic interest in these enzymes, we investigated expression and coregulation in hormone-sensitive (HS) and castrate-resistant (CR) prostate cancer (PC).
Methods
EZH2 and NSD2 levels were quantified using VECTRA analysis in HS and CRPC tissue microarrays (n = 105 + 66). Expression data from The Cancer Genome Atlas (n = 498), Memorial Sloan Kettering Cancer Center (n = 240), and Stand Up to Cancer/Prostate Cancer Foundation (n = 444) cBioportal datasets were queried, and associations between EZH2 and NSD2 and clinicopathologic variables determined.
Results
Tumour expression of NSD2, but not EZH2, increased in CRPC (p = 0.05, 0.09). Epithelial nuclei co-expressing NSD2 and EZH2 increased in CRPC compared to HSPC (69 vs 42%, p = 0.02), and in metastatic tissue relative to benign (55 vs 35%, p = 0.02). cBioportal analysis revealed collinear NSD2/EZH2 expression (Spearman = 0.57, 0.58, 0.58, all p < 0.001). NSD2/EZH2 co-expression significantly associates with clinicopathologic characteristics including grade group, stage and seminal vesicle involvement. On univariate and multivariate analysis tumours co-expressing NSD2 and EZH2 conferred increased risk of recurrence (hazard ratio: 2.6, 95% confidence inerval: 1.2–5.4, p = 0.01). Kaplan–Meier analysis revealed reduced progression-free-survival of NSD2 and EZH2 co-expression patients in datasets (p < 0.001, 0.002).
Conclusions
Increased EZH2/NSD2 co-expression is overrepresented in CRPC, metastases and associates with shorter disease-free survival in PC patients. Coregulation of these two histone methyltransferases is a biomarker for aggressive PC and licenses them as therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Wilson, B. G. & Roberts, C. W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011).
Kouzarides, T. SnapShot: histone-modifying enzymes. Cell 128, 802.e1–02.e2 (2007).
Xu, K., Wu, Z. J., Groner, A. C., He, H. H., Cai, C., Lis, R. T. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012).
Varambally, S., Dhanasekaran, S. M., Zhou, M., Barrette, T. R., Kumar-Sinha, C., Sanda, M. G. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Asangani, I. A., Ateeq, B., Cao, Q., Dodson, L., Pandhi, M., Kunju, L. P. et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol. Cell 49, 80–93 (2013).
Friedman, J. M., Jones, P. A. & Liang, G. The tumor suppressor microRNA-101 becomes an epigenetic player by targeting the polycomb group protein EZH2 in cancer. Cell Cycle 8, 2313–2314 (2009).
Varambally, S., Cao, Q., Mani, R. S., Shankar, S., Wang, X., Ateeq, B. et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 (2008).
Nikoloski, G., Langemeijer, S. M., Kuiper, R. P., Knops, R., Massop, M., Tonnissen, E. R. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42, 665–667 (2010).
Popovic, R., Martinez-Garcia, E., Giannopoulou, E. G., Zhang, Q., Ezponda, T., Shah, M. Y. et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 10, e1004566 (2014).
Li, J., Ahn, J. H. & Wang, G. G. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol. Life Sci. 76, 2899–2916 (2019).
Nimura, K., Ura, K., Shiratori, H., Ikawa, M., Okabe, M., Schwartz, R. J. et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 460, 287–291 (2009).
Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Wang, G., Zhao, D., Spring, D. J. & DePinho, R. A. Genetics and biology of prostate cancer. Genes Dev. 32, 1105–1140 (2018).
Damodaran, S., Damaschke, N., Gawdzik, J., Yang, B., Shi, C., Allen, G. O. et al. Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer 17, 874 (2017).
Lee, J. H., Yang, B., Lindahl, A. J., Damaschke, N., Boersma, M. D., Huang, W. et al. Identifying dysregulated epigenetic enzyme activity in castrate-resistant prostate cancer development. ACS Chem. Biol. 12, 2804–2814 (2017).
Desmeules, P., Hovington, H., Nguile-Makao, M., Leger, C., Caron, A., Lacombe, L. et al. Comparison of digital image analysis and visual scoring of KI-67 in prostate cancer prognosis after prostatectomy. Diagn. Pathol. 10, 67 (2015).
Taylor, B. S., Schultz, N., Hieronymus, H., Gopalan, A., Xiao, Y., Carver, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010).
Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., Lazar, A. J., Cherniack, A. D. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–16.e11 (2018).
Abida, W., Cyrta, J., Heller, G., Prandi, D., Armenia, J., Coleman, I. et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA 116, 11428–11436 (2019).
Aytes, A., Giacobbe, A., Mitrofanova, A., Ruggero, K., Cyrta, J., Arriaga, J. et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat. Commun. 9, 5201 (2018).
Kang, H. B., Choi, Y., Lee, J. M., Choi, K. C., Kim, H. C., Yoo, J. Y. et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 583, 1880–1886 (2009).
Zhang, Y., Zheng, D., Zhou, T., Song, H., Hulsurkar, M., Su, N. et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 9, 4080 (2018).
Bachmann, I. M., Halvorsen, O. J., Collett, K., Stefansson, I. M., Straume, O., Haukaas, S. A. et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 24, 268–273 (2006).
Saramäki, O. R., Tammela, T. L., Martikainen, P. M., Vessella, R. L. & Visakorpi, T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer 45, 639–645 (2006).
Tian, X., Tao, F., Zhang, B., Dong, J. T. & Zhang, Z. The miR-203/SNAI2 axis regulates prostate tumor growth, migration, angiogenesis and stemness potentially by modulating GSK-3β/β-CATENIN signal pathway. IUBMB Life 70, 224–236 (2018).
Saini, S., Majid, S., Yamamura, S., Tabatabai, L., Suh, S. O., Shahryari, V. et al. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin. Cancer Res. 17, 5287–5298 (2011).
Taplin, M.-E., Hussain, A., Shah, S., Shore, N. D., Agrawal, M., Clark, W. et al. ProSTAR: a phase Ib/II study of CPI-1205, a small molecule inhibitor of EZH2, combined with enzalutamide (E) or abiraterone/prednisone (A/P) in patients with metastatic castration-resistant prostate cancer (mCRPC). J. Clin. Oncol. 37(Suppl.), TPS335–TPS335 (2019).
Acknowledgements
We thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory (TRIP), supported by the UW Department of Pathology and Laboratory Medicine, UWCCC (P30 CA014520) and the Office of the Director-NIH (S10OD023526) for use of its facilities and services.
Author information
Authors and Affiliations
Contributions
M.F. organised and interpreted raw data, performed the statistical analysis, designed figures and tables and drafted the manuscript. J.G. was instrumental in designing the experiment and performing data compilation and analysis. A.T. compiled and organised raw data including statistical analysis. G.A. provided statistical analysis and data organisation. W.H. read the H&E slides and was involved in idea development, data development and review, and editing. T.K. provided analysis of raw results and assistance with figures and data presentation. R.M. compiled and analysed raw data involving the tissue staining results. P.L. assisted in idea development, data development and review, and editing of the manuscript. B.Y. assisted in statistical analysis and construction of figures. J.D. was involved in idea development, data development and editing. D.J. designed and supervised the study, interpreted data and edited the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Individual medical centres obtained institutional review board approval exemption or waiver for the use of archived clinical samples for research purposes. This study was performed in accordance with the Declaration of Helsinki. Data and outcomes for cBioportal are made through a data-sharing agreement. Approvals and patient consents were obtained through the University of Wisconsin Carbone Cancer Center Tissue Biobank (IRB #2016-0934) and through the University of Wisconsin Institutional Review Board IRB# XPO5338.
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary information files. RNA expression data from the TCGA, MSKCC and SU2C/PCF is available publicly online from the cBioPortal for cancer genomics.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by the Department of Defense Grant-DODPCRP W81XWH (to D.J.).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Filon, M., Gawdzik, J., Truong, A. et al. Tandem histone methyltransferase upregulation defines a unique aggressive prostate cancer phenotype. Br J Cancer 125, 247–254 (2021). https://doi.org/10.1038/s41416-021-01398-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01398-7