Abstract
In advanced prostate cancer, CREB (cAMP-responsive element-binding protein) binding protein (CBP) and its homolog EP300 are highly expressed; targeting the bromodomain of CBP is a new strategy for the treatment of prostate cancer. In the current study we identified Y08197, a novel 1-(indolizin-3-yl) ethanone derivative, as a selective inhibitor of CBP/EP300 bromodomain and explored its antitumor activity against prostate cancer cell lines in vitro. In the AlphaScreen assay, we demonstrated that Y08197 dose-dependently inhibited the CBP bromodomain with an IC50 value at 100.67 ± 3.30 nM. Y08197 also exhibited high selectivity for CBP/EP300 over other bromodomain-containing proteins. In LNCaP, 22Rv1 and VCaP prostate cancer cells, treatment with Y08197 (1, 5 μM) strongly affected downstream signaling transduction, thus markedly inhibiting the expression of androgen receptor (AR)-regulated genes PSA, KLK2, TMPRSS2, and oncogenes C-MYC and ERG. Notably, Y08197 potently inhibited cell growth in several AR-positive prostate cancer cell lines including LNCaP, 22Rv1, VCaP, and C4-2B. In 22Rv1 prostate cancer cells, treatment with Y08197 (1, 4, 16 μM) dose-dependently induced G0/G1 phase arrest and apoptosis. Furthermore, treatment with Y08197 (5 μM) significantly decreased ERG-induced invasive capacity of 22Rv1 prostate cancer cells detected in wound-healing assay and cell migration assay. Taken together, CBP/EP300 inhibitor Y08197 represents a promising lead compound for development as new therapeutics for the treatment of castration-resistant prostate cancer.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) has the highest number of new cancer cases and the second highest cancer mortality rate in men in the United States; PCa accounted for almost one in five new cancer diagnoses in 2018 [1]. Since androgen receptor (AR) signaling has a pivotal role in PCa, androgen deprivation therapy with surgical or chemical castration is the first option for PCa treatment. Despite the initial benefit, patients often progress to a lethal type of PCa known as castration-resistant prostate cancer (CRPC). Second-generation AR-axis inhibitors, such as the androgen synthesis inhibitor abiraterone acetate (ABI) and the androgen signaling inhibitor enzalutamide (Enz), have been approved by the FDA for metastatic CRPC in 2011 and 2012, respectively. Although both drugs benefit patients and result in improved survival, approximately one-third of patients demonstrate no serum PSA (Prostate specific antigen) response to these drugs [2,3,4]. The remaining patients may acquire secondary resistance, which is marked by a restoration of AR signaling and serum PSA levels [5]. There are multiple mechanisms of resistance, including AR amplification, point mutations (particularly in the AR ligand binding domain (LBD)), and the expression of various AR splice variants, such as AR-V7 [6, 7]. Current therapeutic development is still largely focused on antiandrogens that have increased potencies compared to the potencies of previous therapeutics. Therefore, alternative strategies for the development of new therapies for CRPC treatment are urgently needed.
To overcome the acquired secondary resistance, alternative strategies to tackle AR signaling were developed. Agents disrupting AR N-terminal domain (NTD) functionality without directly targeting the AR LBD or promoting AR degradation are promising solutions that are currently under investigation [8,9,10,11,12,13]. Sadar and others have shown that EPI-001 covalently binds to the AR NTD, blocking the transcriptional activity of AR and AR splice variants and reducing the growth of CRPC xenografts [14, 15]. Raina and others have demonstrated that the small molecule PROTAC inhibitor ARV-771, an AR transcriptional coactivator BET degrader, showed potent suppression of AR signaling, AR levels and tumor growth in a CRPC mouse xenograft model [16]. We have proven that retinoic acid receptor-related orphan receptor γ (RORγ) acts as an upstream determinant of AR gene expression and that RORγ antagonists can potently inhibit the growth of various AR-positive xenograft tumors in mice [17, 18].
Nuclear receptor signaling is dependent on a number of transcriptional coactivators that enhance the binding of the receptor to chromatin and promote the transcriptional activation of target genes [19]. The CREB (cAMP-responsive element-binding protein) binding protein (CBP) and its homolog, EP300, are fundamental coactivators among a large number of transcription factors [20]. Mechanistically, CBP and EP300 interact with the AR NTD to potentially stabilize both the N/C interactions and the binding of AR to AREs (AR response elements) [21]. CBP and EP300 act as steroid receptor coactivators for various nuclear receptors, including the retinoid X receptor (RXR), estrogen receptor (ER) and AR [22,23,24,25], which have been implicated in the development of various cancers, inflammatory conditions, and other diseases [26,27,28,29]. In advanced PCa, CBP and EP300 are highly expressed, and androgen deprivation leads to the upregulation of both proteins [19, 30]. Therefore, interrupting the interaction of AR with its coactivator CBP by targeting the bromodomain of CBP may be an efficient strategy to block AR signaling. The CBP bromodomain may represent a potential therapeutic target for PCa. Recently, we and others have proven that CBP and EP300 bromodomain inhibition with small molecule inhibitors could suppress PCa growth in vitro and in vivo [31, 32]. We have also reported the fragment-based virtual screen and medicinal chemistry optimization of a series of 1-(1H-indol-1-yl)ethanone derivatives as CBP and EP300 bromodomain inhibitors, which inhibited cell growth in several PCa cell lines and reduced the expression of AR, AR-V7 and PSA [32].
In this study, we report that Y08197, a novel 1-(indolizin-3-yl)ethanone derivative optimized from our previous study [32], potently and selectively binds to the CBP bromodomain. Y08197 markedly inhibits cell growth, proliferation and migration in several PCa cell lines. Y08197 also reduces the expression of PSA, KLK2, TMPSS2 and other oncogenes in LNCaP, 22Rv1, and VCaP cells. Y08197 represents a promising lead compound for further optimization. Thus, this study further validates that the disruption of interactions between an AR and its coactivator CBP may be a promising strategy for the treatment of CRPC and other diseases.
Materials and methods
Compounds
Y08197 (3-Acetyl-N-(2-fluoro-5-(hydroxymethyl)-3-(1-methyl-1H-pyrazol-4-yl)phenyl)-7-methoxyindolizine-1-carboxamide) was synthesized by the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (Guangzhou, China). Chemical formula: C23H21FN4O4, MW: 436.44. CPI-637 and enzalutamide were purchased from Selleck.
Protein expression and purification
The bromodomains were expressed as a His6-fusion protein with a TEV (Tobacco Etch Virus) cleavage site between His6 and the bromodomain using the pET24a expression vector (Novagen). The cDNA sequences encoding the bromodomain of human CBP (residues R1081-G1197), EP300 (residues A1040-G1161), BRD4(1) (residues N44-E168), BRD7 (residues E1-R108), PCAF (residues G715-D831), BAZ2B (residues S1858-S1972), TRIM24(1) (residues G896-E1014), TAF1(1) (residues R1377-D1503), ASH1L (residues E2433-E2564), ATAD2A (residues Q981-R1108), and BRPF1 (residues E627-G740) were synthesized by Genscript. The BL21 (DE3) cells that were transformed with these expression plasmids were grown in LB broth at 25 °C to an OD600 of approximately 1.0 and then were induced with 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 16 °C for 16 h. The cells were harvested by centrifugation (6000 × g for 15 min at 4 °C, JLA 81000 rotor, on a Beckman Coulter Avanti J-20 XP centrifuge) and were frozen at −80 °C as pellets for storage. The cells were resuspended in extract buffer (50 mM HEPES (2-[4-(2-Hydroxyethyl)-1-piperazinyl]ethanesulfonic acid), pH 7.5 at 25 °C, 500 mM NaCl, 5 mM imidazole, 5% glycerol, and 0.5 mM TCEP (tris(2-carboxyethyl)phosphine hydrochloride)) and were high-pressure homogenized using a JN3000 PLUS high-pressure homogenizer (JNBIO, Guangzhou, China) at 4 °C. The lysate was collected on ice and was centrifuged at 12 000 × g for 40 min. The supernatant was loaded onto a 5 mL NiSO4-loaded HisTrap HP column (Ni-NTA, GE Healthcare, NJ, USA). The column was washed with 20 mL of extract buffer (50 mM HEPES, pH 7.5 at 25 °C, 500 mM NaCl, 50 mM imidazole). The protein was eluted with a 50–500 mM imidazole gradient in elution buffer (50 mM HEPES, pH 7.5 at 25 °C, 500 mM NaCl, 500 mM imidazole). The protein was concentrated and further purified by a gel filtration column (HiLoad, Superdex 75, 16/60, GE Healthcare, USA). The sample purity of each fraction was examined by SDS-PAGE, and the sample concentration was determined by a Bradford assay. The purified proteins were concentrated in gel filtration buffer (10 mM HEPES pH 7.5 at 25 °C, 150 mM NaCl, 0.5 mM TCEP) and were stored at −80 °C for an AlphaScreen assay or a TSA.
AlphaScreen assay
Interactions between bromodomain-containing proteins (BCP) and ligands were assessed by luminescence-based AlphaScreen technology using a histidine detection kit from PerkinElmer (Norwalk, CT, USA). All of the reactions contained BCP bound to nickel acceptor beads (5 μg/mL) and biotinylated and acetylated histone H4 peptides bound to streptavidin donor beads (5 μg/mL) in the presence or absence of the indicated amounts of Y08197 or CPI-637. The C-terminal biotinylated tetra-acetylated histone peptide H4 (bH4KAc4) sequence was H-SGRGK(Ac)GGK(Ac)GLGK(Ac)GGAK(Ac)RHRK-Biotin-OH (synthesized by Genscript). The experiments were conducted with a protein/peptide ratio for the sensitive signal as follows: CBP:bH4KAc4 = 150 nM:50 nM. All reagents were diluted in buffer (50 mM MOPS (pH 7.4), 50 mM NaF, 50 μM CHAPS, and 0.1 mg/mL bovine serum albumin) and were allowed to equilibrate at room temperature prior to being added into low-volume 384-well plates (ProxiPlate-384 Plus, PerkinElmer, USA). The plates were wrapped in foil to protect the reagents from light. Then, the plates were incubated at room temperature for 2.5 h and were read on an EnSpire plate reader (PerkinElmer, USA). When excited by a laser beam at 680 nm, the donor beam emits singlet oxygen that activates the thioxene derivatives in the acceptor beads, which releases photons of 520–620 nm as the binding signal. All experiments were performed in triplicate on the same plate. The results were based on an average of three experiments, and the standard errors were typically less than 10% of the measurements.
Thermal stability shift assay (TSA)
Thermal stability shift assays were carried out using the Bio-Rad CFX96 Real-Time PCR system. All reactions were buffered in 10 mM HEPES, pH 7.5, 150 mM NaCl at final concentrations of 10 μM for the proteins and 200 μM for the compounds. The 20 μL reaction mixture was added to the wells of a 96-well PCR plate. SYPRO Orange (ABI, Sigma) was added as a fluorescence probe at a dilution of 1:1000 and was incubated with the compounds on ice for 30 min. The total DMSO concentration was restricted to 1% or less. The excitation and emission filters for the SYPRO Orange dye were set to 465 and 590 nm, respectively. The temperature was increased at steps of 0.3 °C per minute from 30 to 75 °C, and the fluorescence readings were taken at each (0.3 °C) interval. All experiments were performed in triplicate. The melting temperatures (Tm) were calculated by fitting the sigmoidal melting curve to the Boltzmann equation using GraphPad Prism. ΔTm is the difference in the Tm values calculated for the reactions with and without compounds.
Cell culture, RNA isolation, and quantitative real-time PCR
LNCaP, 22Rv1, C4-2B, and VCaP PCa cells were cultured in RPMI-1640 containing 10% FBS (Fetal bovine serum) at 37 °C in 5% CO2 incubators. For qRT-PCR analysis, LNCaP, 22Rv1, and VCaP cells were seeded at 1.5 × 105 cells per well in 12-well plates. Twenty-four hours later, the cells were treated with compounds (1 and 5 μM, respectively) for 48 h. The total RNA was then isolated with the Eastep Super Total RNA Extraction Kit (Promega, USA), and the cDNA was synthesized from 1000 ng total RNA using the All-in-oneTM First-Strand cDNA Synthesis Kit (Genecopoeia, USA). qRT-PCR was performed in triplicate using standard SYBR green reagents. The full-length AR (AR-FL), AR-V7, PSA (KLK3), KLK2, TMPRSS2, C-MYC and ERG gene expression levels were assessed by real-time PCR and were normalized to the expression levels of β-actin. The primer sequences for qRT-PCR are listed in Table 1.
Cell lysates and Western blot analysis
The 22Rv1 cells were treated with the vehicle or different concentrations of Y08197, CPI-637 or Enz for 48 h before being harvested for a Western blot analysis. The cells were lysed in RIPA buffer plus 1% PMSF (Phenylmethylsulfonyl fluoride) and then were separated on a denaturing polyacrylamide gel according to the manufacturer’s instructions. The proteins were transferred to a PVDF (Polyvinylidene fluoride) membrane and were blocked for 2 h in a 5% nonfat milk solution (Tris-buffered saline, 0.1% Tween (TBS-T), 5% nonfat dry milk). The membranes were incubated overnight with the following antibodies: AR (Abclonal; A2053), C-MYC (Santa Cruz; sc-40), ERG (Abcam; ab87105), PSA (Biowold; BS6241) and GAPDH (Abclonal; AC001). Then, the membranes were incubated with HRP-linked secondary antibodies (Proteintech, SA00001; Abclonal, AS014). Finally, the signal was detected using a Minichemi imager.
PSA luciferase reporter gene assay
LNCaP cells were seeded into 24-well plates and were transiently transfected using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. LNCaP cells were cotransfected with 200 ng PSA-Luc reporter plasmid and 10 ng Renilla luciferase expression plasmid per well. Chemicals were added 24 h after transfection. The cells were harvested after another 24 h for a luciferase assay using the dual-luciferase reporter assay system (Promega, USA). The luciferase activities were normalized to the Renilla activity, which was cotransfected as an internal control. All of the assays were performed in triplicate, and the standard deviations were calculated accordingly.
Cell viability and cell colony formation assays
For the cell viability assessment, the cells were seeded into 384-well plates at 500–1000 cells per well (optimum density for growth) in a total volume of 20 μL of media. After 12 h, 10 μL of the appropriate chemical compound with a twofold serial dilution was added to a final volume of 30 μL of media to each well with a final concentration of 5 nM to 100 μM. The measurement was conducted 96 h after seeding the LNCaP, C4-2B, 22Rv1, PC-3 cells, 144 h after seeding the VCaP cells and 72 h after seeding the Du145 cells. Then, 25 μL of Cell-Titer GLO reagents (Promega, USA) was added, and the luminescence was measured on a GLOMAX Microplate Luminometer (Promega, USA) according to the manufacturer’s instructions. The estimated in vitro half-maximal inhibitory concentration (IC50) values were calculated using GraphPad Prism 6 software.
For the C4-2B and 22Rv1 colony formation assay, 2000 cells per well were seeded into a six-well plate. The cells were cultured with the vehicle or the indicated concentrations of compounds for 14 d with 3 mL medium. When the cell colony was first visible, the medium was removed, and the plates were washed once with 2 mL PBS (Phosophate-buffered saline). The cell colonies were stained with 2.5% crystal violet (in MeOH) for 2 h. The plates were scanned with an HP scanner.
Cell cycle determination
22Rv1 cells were collected and dispersed into six-well plates with 3 × 105 cells per well. The different concentrations of Y08197, CPI-637 and Enz were used to treat the cells for 48 h. The cells were collected and fixed with 70% ethanol overnight. After washing twice with PBS, the cells were resuspended in 500 µL of PBS containing 50 µg/mL propidium iodide (PI) and 100 μg/mL RNase A for 60 min at 20 °C in the dark. The flow cytometry analysis was performed using a BD LSR Fortessa SORP Flow Cytometer (BD Biosciences, San Jose, CA, USA), and the data were analyzed using Modfit LT 5.0 software.
Cell apoptosis examination
The levels of apoptosis were investigated using the Annexin V-FITC Apoptosis Detection Kit (KeyGEN; KGA106). 22Rv1 cells were seeded into a six-well plate and were incubated in gradient concentrations of Y08197, CPI-637, and Enz for 48 h. The cells were washed twice with PBS and were resuspended in 500 µL of PBS containing 5 μL of Annexin V-FITC and 5 µL of the PI Staining Solution for 15 min. The flow cytometry analysis was performed using a BD LSR Fortessa SORP Flow Cytometer (BD Biosciences, San Jose, CA, USA), and the data were analyzed using FlowJo_V10 software.
Wound-healing assay
22Rv1 cells were seeded into a 12-well plate and were allowed to grow to nearly 100% confluence in the culture medium. Subsequently, a cell-free line was manually drawn by scratching the confluent cell monolayer with a 10 μL pipet tip. The wounded cell monolayer was washed three times with PBS and was incubated in RPMI-1640 (containing 2% FBS) with the vehicle, 5 μM Y08197, CPI-637 or Enz for 48 h. One scratched field was chosen at random, and the images were captured with a bright-field microscope (CKX41; Olympus). The percentage of wound closure was measured using Adobe Photoshop 7.0.1 (Adobe Systems Inc., San Jose, CA, USA).
Cell migration assay
Cell migration assays were evaluated in 8.0 mm Transwell chambers (Corning Costar, USA). A total of 5 × 104 22Rv1 cells with either the vehicle, 5 μM Y08197, CPI-637 or Enz were plated in the top chamber with medium that did not contain FBS. Complete media was added to the lower chamber as a chemoattractant. After 48 h of incubation at 37 °C in 5% CO2, the nonmigrating cells were gently removed with a cotton swab. The migrated cells that were located on the lower side of the membranes were fixed with 4% formaldehyde and were stained with 0.25% crystal violet. The migrated cells were quantitated by counting the cells and were graphed in three randomly selected fields. The images were captured with a bright-field microscope (CKX41; Olympus).
Statistical analysis
Numerical data from at least three individual experiments are shown as the mean ± SD unless otherwise indicated. Statistical significance was assessed by using unpaired two-tailed Student’s t test. In statistical comparisons, a P value < 0.05 was used to indicate a statistically significant difference except the data of the PSA-luciferase reporter assay.
Results
Y08197 is a potent and selective CBP bromodomain inhibitor
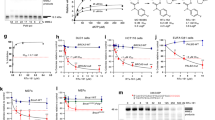
The inhibition of Y08197 (Fig. 1a) against the recognition of acetylated histones by the CBP bromodomain was evaluated using an AlphaScreen assay (Fig. 1b). The histidine-tagged CBP bromodomain was immobilized on Ni-coated acceptor beads, and the biotinylated H4Kac4 peptide was immobilized on streptavidin-coated donor beads. The donor beads contained a photosensitizer that converts ambient oxygen into singlet oxygen upon activation with light at 680 nm. If the acceptor beads are brought within close proximity of the donor beads by a bromodomain-acetylated lysine interaction, energy is transferred from the singlet oxygen to the thioxene derivatives in the acceptor beads, and this results in light emission at 520–620 nm. The addition of a CBP-bromodomain inhibitor blocks the signal conduction by acceptor bead-immobilized CBP and by donor bead-immobilized H4Kac4. Y08197 potently and dose-dependently inhibited the CBP bromodomain with an IC50 value of 100.67 ± 3.30 nM (Fig. 1c). Since the structures of the bromodomain pockets are highly similar, we also investigated the selectivity profile of Y08197 for the CBP and EP300 bromodomains over those from other proteins from different subgroups of the bromodomain family. Using a thermal stability shift assay (TSA), the abilities of Y08197 and CPI-637 to maintain stability on 11 BCPs were profiled. The results demonstrate that Y08197 exhibited potent inhibition only on the CBP and EP300 bromodomains with minimal inhibition activity on the remaining tested proteins (Fig. 1d). However, CPI-637 showed slight additional inhibitory activity on BRD4 and BRD7 in addition to CBP and EP300 (Fig. 1d). These findings suggest that Y08197 is a potent and selective inhibitor of the CBP and EP300 bromodomains in vitro.
Y08197 is a selective CBP bromodomain inhibitor. a The chemical structures of Y08197, CPI-637 and Enz. b The schematic representation of the AlphaScreen (Amplified Luminescent Proximity Homogeneous Assay Screen) assay. c Y08197 and CPI-637 dose-dependently inhibited the CBP bromodomain and acetylated histone H4 peptide interactions as determined by AlphaScreen assays. The IC50 was calculated from the AlphaScreen assay. The data are expressed as the means ± SD and are represented from at least three independent experiments. d The selectivity of Y08197 and CPI-637 against bromodomain-containing proteins. The bromodomain selectivity profiles were determined by a thermal shift assay. Compound concentration, 200 μM; protein concentration, 10 μM
The CBP inhibitor Y08197 inhibits AR signaling in prostate cancer cells
To better understand how Y08197 inhibits the expression of AR, AR-V7, AR-regulated genes and some oncogenes, a qRT-PCR analysis was performed in LNCaP, 22Rv1 and VCaP cells with the treatment concentrations of 1 and 5 μM, respectively. As shown in Fig. 2b, c, the AR-regulated genes PSA (KLK3), KLK2, and TMPRSS2 were significantly suppressed at the mRNA level upon Y08197 treatment compared to Enz in 22Rv1 and VCaP cells. CPI-637, a CBP inhibitor from Constellation Pharmaceuticals, exhibited a similar inhibitory activity as that of Y08197. As indicated by the qRT-PCR assays shown in Fig. 2, the C-MYC mRNA level was suppressed upon Y08197 treatment, especially in LNCaP and 22Rv1 cells. ERG is a potential PCa therapeutic target, and its transcription factor product is aberrantly upregulated in PCa due to chromosomal rearrangements between the androgen-regulated genes TMPRSS2 and ERG [33,34,35]. Here, a qRT-PCR analysis (Fig. 2c) showed that the ERG mRNA levels were also suppressed upon Y08197 treatment in VCaP cells. In addition, the CBP bromodomain inhibitors Y08197 and CPI-637 can also suppress the mRNA expression of full-length AR (AR-FL) transcripts in LNCaP cells and can suppress the expression of AR-V7 in 22Rv1 cells; no obvious activity was observed upon AR inhibitor Enz treatment.
The CBP inhibitor Y08197 inhibits AR signaling at mRNA level in prostate cancer cells. qRT-PCR analysis of indicated genes in LNCaP (a), 22Rv1 (b) and VCaP (c) cells treated with 1 or 5 μM of Enz, CPI-637 or Y08197 for 2 d. The data are expressed as the mean ± SEM (n = 3). *P < 0.05, **P < 0.01 by two-tailed Student’s t test
Furthermore, a Western blot analysis was performed to analyze how Y08197 influenced the protein expression status in 22Rv1 cells. As shown in Fig. 3a, Y08197 has a modest inhibitory activity towards the protein expression of AR-FL and AR-V7 at 1 and 4 μM, respectively, which is consistent with the mRNA expression inhibition shown in Fig. 2. For C-MYC, Y08197 displayed a significant dose-dependent inhibitory activity similar to that of CPI-637, which suppressed nearly 50% of the protein expression when the treatment concentration was reduced to a relatively low level at 1 μM. Figure 3a shows that Y08197 exhibited no inhibitory activity towards the protein expression of ERG until the treatment concentration was increased to 8 μM.
The CBP inhibitor Y08197 inhibits AR signaling at transcription and protein levels in prostate cancer cells. a Western blot analysis of AR-FL, AR-VS, C-MYC, ERG, PSA in 22Rv1 cells treated with Enz, CPI-637 or Y08197 for 2 d. GAPDH served as the loading control. b PSA transcriptional activity was evaluated by luciferase reporter assays in LNCaP cells transfected with PSA-luc reporter plasmid. The cells were treated with the vehicle or with 0.4, 2.0 or 10.0 μM of Enz, CPI-637 or Y08197. The data are expressed as the mean ± SEM (n = 3). *P < 0.01, **P < 0.001, ***P < 0.0001 by two-tailed Student’s t test
As PSA is currently a well-recognized biomarker for PCa, in addition to PSA mRNA and protein expression, a PSA promoter-driven luciferase reporter assay was also designed as a surrogate to further evaluate the potential impact of Y08197 on PSA transcription. Y08197 dose-dependently inhibited the PSA reporter activity and displayed a comparable inhibitory effect to that of CPI-637 on the mRNA, transcription and protein levels (Figs. 2, 3). The results suggest that the CBP bromodomain inhibitor Y08197 could effectively influence related gene expression and could inhibit AR signaling in AR-positive PCa cells.
The CBP inhibitor Y08197 inhibits prostate cancer cell line proliferation
To investigate the cellular antiproliferation activities, we performed cell viability assays in the AR-positive PCa cell lines LNCaP, C4-2B, 22Rv1 and VCaP and in the AR-negative PCa cell lines Du145 and PC-3 (Fig. 4). The second-generation androgen receptor antagonist Enz and the CBP bromodomain inhibitor CPI-637 were used as references. The results indicate that the CBP bromodomain inhibitor Y08197 inhibited AR-positive PCa cell growth with higher potency than that exhibited by Enz (Fig. 4). In AR-negative PCa cell lines, Y08197 showed a minimum inhibitory rate in PC-3 and Du145 cells (Fig. 4).
The CBP inhibitor Y08197 inhibits prostate cancer cell line proliferation. a Cell viability was measured by Cell-Titer GLO for the LNCaP, C4-2B, 22Rv1, VCaP, Du145, PC-3 prostate cancer cells treated with the indicated concentrations of Enz, CPI-637 or Y08197. The experiments were performed in triplicate. b The half-maximum inhibitory concentration (IC50) values for all compounds in each cell line are shown
To further investigate the long-term growth inhibition effects, colony formation assays with C4-2B and 22Rv1 cells were performed for the CBP inhibitors Y08197 and CPI-637 and the AR antagonist Enz (Fig. 5). Similar to the cell viability assay, Enz showed no inhibitory activity for 22Rv1 cell colony formation under the treatment concentration range. The Y08197 compound reduced the colony formation abilities of C4-2B and 22Rv1 in a dose-dependent manner. The colony formation was reduced to less than 10% in 22Rv1 cells at 16 μM and in C4-2B cells at 4 μM. Consistent with the cell viability assay data, the Y08197 compound showed a moderate antiproliferation activity.
As CBP and EP300 have been investigated to be involved in the development of various diseases, the antiproliferation activity of Y08197 was also evaluated against a wide range of cancer cell lines (Supplementary Fig. S1). Y08197 displayed approximately twofold more potent inhibitory activities than those of CPI-637 against the breast cancer cell lines MCF-7 (ER positive) and Hs578T (triple negative), with IC50 values of 10.61 and 4.02 μМ, respectively. Y08197 also showed obvious antiproliferation activity against HT-29 human colon cancer cells and HeLa cervical cancer cells with IC50 values of 2.12 and 12 μM, respectively. However, no significant inhibition by Y08197 was observed in the osteosarcoma cell line U2OS, the non-small-cell lung cancer cell line A549 or the lung cancer cell line H1975. The growth inhibitory activities of the compound against lung fibroblast cells HFL-1 were also evaluated to monitor the potential toxic effects. As shown in Supplementary Fig. S1, Y08197 exhibited lower cytotoxic effects than the effects of CPI-637 against normal cells and had a suppression level of 50%, even when the concentration was as high as 100 μM; this indicates that the cytotoxic effect of Y08197 was minimal.
The CBP inhibitor Y08197 induces G1 phase arrest and apoptosis in 22Rv1 cells
MYC is a transcription factor that regulates the global gene expression through heterodimerization with the protein myc-associated factor X (MAX) [36, 37]. Through the genome-wide regulation of gene expression, MYC was identified to be related to diverse cellular processes, including cell proliferation, cell cycle, apoptosis and metabolism [38,39,40].
Considering the significant suppression of Y08197 on the expression of C-MYC, we investigated the mechanisms underlying the cell proliferation inhibition induced by Y08197 in PCa. For this, 22Rv1 cells were exposed to different concentrations of Y08197 (0, 1, 4, 16 μM) for 96 h. Then, the cell cycle distribution was determined by flow cytometry. The effect of CPI-637 and Enz on cell cycle progression in 22Rv1 cells was also investigated. After 96 h treatment in 22Rv1 cells, Y08197 significantly arrested cell cycle progression at the G1 phase (Fig. 6). Exposure to 16 μM of Y08197 arrested 76.34% of the cells in G1 phase compared with 62.40% of the control cells (Fig. 6a). However, Enz showed no cell cycle progression arrest activity under the treatment conditions we set (Fig. 6b).
The CBP inhibitor Y08197 induces G1 phase arrest in 22Rv1 cells. a The effects of Enz, CPI-637 or Y08197 on 22Rv1 cell cycle distribution. 22Rv1 cells were treated with Enz, CPI-637 or Y08197 at the indicated concentrations for 96 h. The cell cycle distribution was determined by LSRFortessa after propidium iodide staining and was analyzed with ModFitLT. The quantification results are presented (b). The bars represent the mean ± SD
We then proceeded to test the effect of Y08197 on apoptosis in 22Rv1 cells. As illustrated in Fig. 7, Y08197 triggered dose-dependent apoptosis in 22Rv1 cells at a concentration of 16 μM; the ratio of apoptosis in 22Rv1 cells was 6.77% (Fig. 7a) compared with 3.94% in the untreated control cells. Consistent with the results of the cell cycle arrest, Enz still exhibited no apoptosis induction activity under the treatment conditions (Fig. 7b). These results demonstrate that Y08197, a new inhibitor of the CBP bromodomain, induced G1 phase cell cycle arrest and apoptosis, which suppressed the proliferation of 22Rv1 cells.
The CBP inhibitor Y08197 induces apoptosis in 22Rv1 cells. a Enz, CPI-637 or Y08197 induced apoptosis in 22Rv1 cells. The cells were treated with Enz, CPI-637 or Y08197 for 96 h, and the percentages of cells in different apoptosis phases were determined by LSRFortessa and were analyzed with Flowjo VX. The quantification results are presented (b). The bars represent the mean ± SD
The CBP inhibitor Y08197 decreases the invasive capacity of 22Rv1 cells
The most prevalent PCa gene fusion joins the 5′ untranslated region of an androgen-regulated gene, TMPRSS2, with the members of the E-twenty six (ETS) transcription factor family (ERG or ETV1) through frequent chromosomal rearrangements [41]. Such ERG gene fusions occur during the initiation of PCa progression, which can then lead to the transition from high-grade prostatic intraepithelial neoplasia lesions to invasive carcinoma [42, 43]. The TMPRSS2-ERG fusion product was shown to drive a unique transcriptional program, inducing DNA damage, and to promote invasion and metastasis [44].
Encouraged by the promising suppression of ERG gene expression by Y08197 that was seen in the qRT-PCR and Western blot analysis, the migration inhibitory effect of Y08197 on 22Rv1 PCa cells (ERG-positive line) was initially investigated with the well-established wound-healing assay. Compared with the untreated control, treatment with Y08197 inhibited the migration of 22Rv1 cells, which retarded the wound-healing speed by 15.75% (Fig. 8b). The migration inhibitory potency of Y08197 in 22Rv1 PCa cells was further evaluated by a Transwell migration assay. 22Rv1 cells were treated with 5 μM Y08197 for 48 h, and the number of cells that migrated to the other side of the filter membrane was quantified. It was shown that Y08197 attenuated the migration of 22Rv1 cancer cells by 18.7% at 5 μM (Fig. 8d). Together, these data demonstrate that Y08197 is able to decrease ERG-induced cell invasive capacity.
The CBP inhibitor Y08197 decreases the invasive capacity of 22Rv1 cells. a Photomicrographs from the scratch-motility assay of 22Rv1 cells treated with 5 μM of Enz, CPI-637 or Y08197 for 2 d. b The quantitative analysis of wound healing. c Photomicrographs from the Transwell migration assay of 22Rv1 cells treated with 5 μM of Enz, CPI-637 or Y08197 for 2 d. d The quantitative analysis of migration. The data are expressed as the mean ± SD (n = 3). *P < 0.05, **P < 0.01 by two-tailed Student’s t test
Discussion
Histone modifications, including acetylation and deacetylation, are the major driving force for epigenetic gene regulation [45] and have been shown to be important in the dysregulation of genes involved in all of the hallmarks of cancer [46]. Histone acetylation marks are written by histone acetyltransferases (HATs), read by bromodomains (BRDs), and erased by histone deacetylases (HDACs) [47, 48]. Several inhibitors of histone deacetylases have been approved for clinical application in cancer treatment [49]. The enzymes mediating histone modification represent new generation targets for small molecule intervention.
Prostate cancer is the most common malignancy and is the second leading cause of cancer mortality among men. In PCa, the AR transcriptional coactivators CBP and EP300, bearing both the HAT domain and the bromodomain, are highly upregulated and may induce the transcription of AR-responsive genes [50]. Therefore, inhibiting AR signaling by blocking the CBP/EP300 bromodomain should inhibit resistance to current therapies and could have a significant potential for the treatment of PCa. Recently, a few selective CBP bromodomain inhibitors, such as SGC-CBP30, I-CBP112 and GNE-049, have been reported to have potent antitumor activity in vitro and in vivo. However, most of these CBP bromodomain inhibitors have been investigated for use in the treatment of hematological malignancies, and none of them have progressed to clinical development.
Based on this, the aim of this study was to design and identify CBP/EP300 bromodomain inhibitors with a novel scaffold for the treatment of CRPC. We rationally designed and synthesized Y08197, a 1-(indolizin-3-yl)ethanone derivative, as a CBP/EP300 bromodomain inhibitor through structure-based optimization. Our data show that Y08197 dose-dependently inhibited the CBP bromodomain activity, with an IC50 value of 100 nM. Y08197 showed excellent selectivity for the CBP and the EP300 bromodomain over the other nine BCPs from different subgroups of the bromodomain family. These results indicate that Y08197 is a potent and selective inhibitor of the CBP and the EP300 bromodomain and deserves further evaluation.
Since CBP and EP300 act as coactivators of AR signaling, their inhibitor Y08197 strongly affected AR downstream signaling transduction. The inhibition of AR and AR-V7 was modest in 22Rv1 cells at both the mRNA and protein expression levels. Y08197 efficiently inhibited the mRNA expression of the AR-regulated genes PSA, KLK2, TMPRSS2, and the oncogenes C-MYC and ERG. The protein expression results confirmed that Y08197 dose-dependently suppressed the expression of the oncogenes C-MYC and ERG, indicating the induction of cell cycle arrest, apoptosis and migration incapacitation. However, Y08197 showed no obvious inhibition of PSA protein expression in 22Rv1 cells, which was consistent with the result of CPI-637. These results suggested that Y08197 exerted great influence on AR downstream signaling in different PCa cell lines and may strongly inhibit PCa cell proliferation and migration.
Y08197 exhibited potent antiproliferation effects on AR-positive PCa cell lines. Colony formation assays showed that Y08197 was able to inhibit PCa cell proliferation for a relatively long term. Because CPI-637 is also a CBP bromodomain inhibitor, its efficacy on PCa growth has been compared accordingly. Our data suggest that Y08197 and CPI-637 exhibit similar potencies and comparable pharmacological efficacies in PCa. As CBP and EP300 have been reported to be involved in the development of diverse diseases, we also evaluated Y08197 on a panel of various cancer cell lines as well as human normal cells. The results indicate that Y08197 potentially inhibited the human colon cancer cell line HT-29 and some other cancer cells with high sensitivity, while the cytotoxic effect towards the lung fibroblast cell line HFL-1 was minimal. Mechanistic insights have shown that such CBP bromodomain inhibition, regardless of Y08197 or CPI-637, led to the induction of G0/G1 phase arrest rather than G2/M phase arrest as well as apoptosis in a dose-dependent manner. We further noted that Y08197 and CPI-637 could inhibit cell migration; this indicates the potential beneficial effects of Y08197 and CPI-637 on PCa metastasis.
In summary, these findings further validate that the modulation of CBP activity with small molecules represents a promising strategy for the treatment of CRPC and other diseases. The CBP bromodomain inhibitor Y08197 represents a promising starting point for further optimization and may ultimately provide a new class of therapeutics for the treatment of CRPC. It will also be interesting to examine combination therapies involving Y08197 with antiandrogens, such as Enz.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005.
Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97.
Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–48.
Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38.
Karantanos T, Evans CP, Tombal B, Thompson TC, Montironi R, Isaacs WB. Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level. Eur Urol. 2015;67:470–9.
Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–11.
Mostaghel EA, Plymate SR, Montgomery B. Molecular pathways: targeting resistance in the androgen receptor for therapeutic benefit. Clin Cancer Res. 2014;20:791–8.
Myung JK, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013;123:2948–60.
Lai KP, Huang CK, Chang YJ, Chung CY, Yamashita S, Li L, et al. New therapeutic approach to suppress castration-resistant prostate cancer using ASC-J9 via targeting androgen receptor in selective prostate cells. Am J Pathol. 2013;182:460–73.
Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, et al. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20:3198–210.
Yu Z, Cai C, Gao S, Simon NI, Shen HC, Balk SP. Galeterone prevents androgen receptor binding to chromatin and enhances degradation of mutant androgen receptor. Clin Cancer Res. 2014;20:4075–85.
Malik R, Khan AP, Asangani IA, Cieslik M, Prensner JR, Wang X, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med. 2015;21:344–52.
Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–46.
Brand LJ, Olson ME, Ravindranathan P, Guo H, Kempema AM, Andrews TE, et al. EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget. 2015;6:3811–24.
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113:7124–9.
Wang J, Zou JX, Xue X, Cai D, Zhang Y, Duan Z, et al. ROR-gamma drives androgen receptor expression and represents a therapeutic target in castration-resistant prostate cancer. Nat Med. 2016;22:488–96.
Zhang Y, Xue X, Jin X, Song Y, Li J, Luo X, et al. Discovery of 2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide derivatives as new RORgamma inhibitors using virtual screening, synthesis and biological evaluation. Eur J Med Chem. 2014;78:431–41.
Dasgupta S, Lonard DM, O'Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279–92.
Takeyama K, Ito S, Sawatsubashi S, Shirode Y, Yamamoto A, Suzuki E, et al. A novel genetic system for analysis of co-activators for the N-terminal transactivation function domain of the human androgen receptor. Biosci Biotechnol Biochem. 2004;68:1209–15.
Comuzzi B, Nemes C, Schmidt S, Jasarevic Z, Lodde M, Pycha A, et al. The androgen receptor co-activator CBP is up-regulated following androgen withdrawal and is highly expressed in advanced prostate cancer. J Pathol. 2004;204:159–66.
Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103.
Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–73.
Missero C, Calautti E, Eckner R, Chin J, Tsai LH, Livingston DM, et al. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc Natl Acad Sci USA. 1995;92:5451–5.
Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, et al. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–5.
Chekler EL, Pellegrino JA, Lanz TA, Denny RA, Flick AC, Coe J, et al. Transcriptional profiling of a selective CREB binding protein bromodomain inhibitor highlights therapeutic opportunities. Chem Biol. 2015;22:1588–96.
Conery AR, Centore RC, Neiss A, Keller PJ, Joshi S, Spillane KL, et al. Bromodomain inhibition of the transcriptional coactivators CBP/EP300 as a therapeutic strategy to target the IRF4 network in multiple myeloma. Elife. 2016;5:e10483.
Ghosh S, Taylor A, Chin M, Huang HR, Conery AR, Mertz JA, et al. Regulatory T cell modulation by CBP/EP300 bromodomain inhibition. J Biol Chem. 2016;291:13014–27.
Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–31.
Heemers HV, Sebo TJ, Debes JD, Regan KM, Raclaw KA, Murphy LM, et al. Androgen deprivation increases p300 expression in prostate cancer cells. Cancer Res. 2007;67:3422–30.
Jin L, Garcia J, Chan E, de la Cruz C, Segal E, Merchant M, et al. Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration-resistant prostate cancer. Cancer Res. 2017;77:5564–75.
Xiang Q, Wang C, Zhang Y, Xue X, Song M, Zhang C, et al. Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer. Eur J Med Chem. 2018;147:238–52.
Chatterjee P, Choudhary GS, Alswillah T, Xiong X, Heston WD, Magi-Galluzzi C, et al. The TMPRSS2-ERG gene fusion blocks XRCC4-mediated nonhomologous end-joining repair and radiosensitizes prostate cancer cells to PARP inhibition. Mol Cancer Ther. 2015;14:1896–906.
Font-Tello A, Juanpere N, de Muga S, Lorenzo M, Lorente JA, Fumado L, et al. Association of ERG and TMPRSS2-ERG with grade, stage, and prognosis of prostate cancer is dependent on their expression levels. Prostate. 2015;75:1216–26.
Xue X, Zhang Y, Wang C, Zhang M, Xiang Q, Wang J, et al. Benzoxazinone-containing 3,5-dimethylisoxazole derivatives as BET bromodomain inhibitors for treatment of castration-resistant prostate cancer. Eur J Med Chem. 2018;152:542–59.
Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–7.
Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67.
Campisi J, Gray HE, Pardee AB, Dean M, Sonenshein GE. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984;36:241–7.
Calabretta B, Kaczmarek L, Ming PM, Au F, Ming SC. Expression of c-myc and other cell cycle-dependent genes in human colon neoplasia. Cancer Res. 1985;45:6000–4.
Kim T, Cui R, Jeon YJ, Fadda P, Alder H, Croce CM. MYC-repressed long noncoding RNAs antagonize MYC-induced cell proliferation and cell cycle progression. Oncotarget. 2015;6:18780–9.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8.
Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–8.
Park K, Dalton JT, Narayanan R, Barbieri CE, Hancock ML, Bostwick DG, et al. TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J Clin Oncol. 2014;32:206–11.
Wang X, Qiao Y, Asangani IA, Ateeq B, Poliakov A, Cieslik M, et al. Development of peptidomimetic inhibitors of the ERG gene fusion product in prostate cancer. Cancer Cell. 2017;31:532–48 e7.
Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51.
Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92.
Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762.
Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111.
De Souza C, Chatterji BP. HDAC inhibitors as novel anti-cancer therapeutics. Recent Pat Anticancer Drug Discov. 2015;10:145–62.
Zhong J, Ding L, Bohrer LR, Pan Y, Liu P, Zhang J, et al. P300 acetyltransferase regulates androgen receptor degradation and PTEN-deficient prostate tumorigenesis. Cancer Res. 2014;74:1870–80.
Acknowledgements
We gratefully acknowledge financial support from the National Natural Science Foundation of China (grants 81673357 and 21602222), the Natural Science Foundation of Guangdong Province (grant 2015A030312014), the “Personalized Medicines—Molecular Signature-based Drug Discovery and Development”, Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA12020363), the Chinese National Programs for Key Research and Development (grant 2016YFB0201701), Guangdong Provincial Key Laboratory of Biocomputing (grant 2016B030301007) and Guangzhou Regenerative Medicine and Health Guangdong Laboratory (grant 2018GZR110105016). The authors also gratefully acknowledge support from the Guangzhou Branch of the Supercomputing Center of Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
YX, YZ, QPX and LJZ designed the study. LJZ, QPX, XQX, CZ, CCL, CW, QL, RW, and SW performed and analyzed the experiments. LJZ and QPX wrote the manuscript. YX, YZ, YLZ and QPX revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Zou, Lj., Xiang, Qp., Xue, Xq. et al. Y08197 is a novel and selective CBP/EP300 bromodomain inhibitor for the treatment of prostate cancer. Acta Pharmacol Sin 40, 1436–1447 (2019). https://doi.org/10.1038/s41401-019-0237-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-019-0237-5
Keywords
This article is cited by
-
Targeting bromodomain-containing proteins: research advances of drug discovery
Molecular Biomedicine (2023)
-
Transcriptional co-activators: emerging roles in signaling pathways and potential therapeutic targets for diseases
Signal Transduction and Targeted Therapy (2023)
-
Update and Potential Opportunities in CBP [Cyclic Adenosine Monophosphate (cAMP) Response Element-Binding Protein (CREB)-Binding Protein] Research Using Computational Techniques
The Protein Journal (2021)
-
CREB-binding protein (CREBBP) and preeclampsia: a new promising target gene
Molecular Biology Reports (2021)