Abstract
Coronavirus disease 2019 (COVID-19), a highly infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected more than 235 million individuals and led to more than 4.8 million deaths worldwide as of October 5 2021. Cryo-electron microscopy and topology show that the SARS-CoV-2 genome encodes lots of highly glycosylated proteins, such as spike (S), envelope (E), membrane (M), and ORF3a proteins, which are responsible for host recognition, penetration, binding, recycling and pathogenesis. Here we reviewed the detections, substrates, biological functions of the glycosylation in SARS-CoV-2 proteins as well as the human receptor ACE2, and also summarized the approved and undergoing SARS-CoV-2 therapeutics associated with glycosylation. This review may not only broad the understanding of viral glycobiology, but also provide key clues for the development of new preventive and therapeutic methodologies against SARS-CoV-2 and its variants.
Similar content being viewed by others
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus causing the coronavirus disease 2019 (COVID-19), which threatens human health and public safety.1,2,3,4,5 SARS-CoV-2 virus is genetically closely related to SARS-CoV,6,7,8 less deadly but far more transmissible.9,10,11 It usually causes a lower respiratory tract infection, and the most common symptoms include fever, malaise, dry cough and shortness of breath, which can progress to severe acute respiratory syndrome and even multiple organ failure.12,13,14,15,16,17 Epidemiology data show that the SARS-CoV-2 pandemic has resulted in more than 235 million confirmed infected cases and more than 4.8 million deaths worldwide as of October 5 2021 (https://covid19.who.int/), urgently calling for effective prevention and intervention therapeutics.18,19,20 In-depth studies on viral infection and pathogenic mechanisms will help to find potential cures for COVID-19.21,22,23,24
Protein glycosylation is a process of post-translational or co-translational covalent attachment of glycans to the amino acid side chains of proteins.25,26,27 Glycans, being linear or branched chains of monosaccharides, often have high solubility and conformational entropy, which regulates the protein folding, structures, and functions.28,29,30,31,32 The SARS-CoV-2 is decorated by a large number of highly glycosylated proteins,33 and its glycosylation (both N-linked and O-linked) extensively affects host recognition,32,34,35 penetration,36 binding,2 recycling,37 and pathogenesis.38,39,40,41,42 In this review, we have systematically introduced the methods for characterizing protein glycosylation, summarized the reported glycosylome of SARS-CoV-2 proteins and its receptor protein ACE2, described the potential biological functions of the glycosylation in SARS-CoV-2, and presented the approved and potential SARS-CoV-2 prevention and treatment theraputics associated with glycosylation.
Overview of protein glycosylation
Glycosylation is the most common protein post-translational modification (PTM) in virus.43,44,45,46,47 Glycosylation not only promotes viral protein folding and subsequent trafficking,45,48,49 but also modulates their interactions with receptors and the following innate and adaptive immune response,50,51,52 which affects the host recognition, viral replication, and infectivity.53,54,55 The viruses choose the host cell biosynthetic pathway to produce their genetic and structural materials, and thus the glycosylation of viral proteins greatly depends on the host organelles and enzymes.45,47,56,57 As the evolution of viruses, their glycosylome changes, which may cause huge impacts on the survival and transmissibility of the viruses.45
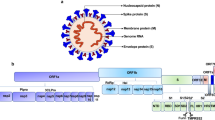
N-glycosylation refers to the glycans attached to asparagine (Asn) residue.43,58 The glycan precursor (Glc3Man9GlcNAc2) containing three glucose (Glc), nine mannose (Man), and two N-acetylglucosamine (GlcNAc) is first synthesized in the membrane of endoplasmatic reticulum (ER).59,60 Then, the glycan precursor is transported to the ER lumen for processing by adding monosaccharides.61 When the glycan is matured, it is added to Asn residue by the oligosaccharyltransferase (OST), and the nascent protein is formed.47 Next, other enzymes like mannosidases, glucosidases, sialyl-, fucosyl-, or galactosyl-transferases located at the ER-Golgi apparatus decorate the protein.50,61 N-linked glycans mainly simplifies into three types based on the structures, including oligomannose (2HexNAc), hybrid (3HexNAc), and complex-type (with more than 3HexNAc) N-glycan structures62,63 (Fig. 1a).
The formation process of N-glycosylation and O-glycosylation in SARS-CoV-2. According to the complexity of the glycans, the N-glycosylation (a) is classified into oligomannose-type (2HexNAc), hybrid-type (3HexNAc), and complex-type (with more than 3HexNAc) glycans, whereas the mucin O-glycosylation (b) is classified into 8 Core types
O-glycosylation usually occurs on serine (Ser), threonine (Thr), and tyrosine (Tyr) residues,43,64 and mucin-type O-glycosylation (N-acetylgalactosamine (GalNAc)-type) is most common in virus.29,65,66 In the O-glycosylation process, GalNAc monosaccharide is first transferred by GalNAc-transferases to Ser, Thr, or Tyr residue in the Golgi apparatus.56,60 The glycosyltransferases then decorate the O-linked glycans, of which eight core structures have been described (Fig. 1b). Core 1-4 are four common O-GalNAc glycan core structures in mammals,67,68 while Core 1 and Core 2 prefer to exist in virus.69,70,71
Methods to characterize protein glycosylation
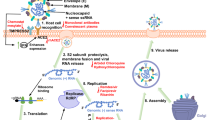
Mass spectrometry (MS)-based N-glycoproteomics has been widely adopted for both site- and structure-specific characterization of glycosylation.2,12,72,73,74,75,76,77,78,79,80 Sample preparation, chromatographic separation, LC-MS/MS analysis and bioinformatics data analysis are the four key pipeline steps.81,82,83,84,85,86,87,88,89,90,91,92
Sample preparation (Fig. 2, left). Glycosylation analysis usually includes the characterization of glycan,93,94,95 intact glycopeptide,96,97,98 glycosite-containing peptide,99,100 as well as intact glycoprotein.101,102,103,104,105 During the sample preparation, glycan releasing enzymes (such as PNGase F for N-glycans),106,107,108,109,110,111 protease (such as trypsin),112,113,114,115,116,117 both glycan releasing and protease,118,119 or no enzymatic approaches may be adopted.120,121,122,123 Glycans usually need to be enriched by hydrophilic materials, such as porous graphitized carbon (PGC),124,125 before MS analysis,126,127,128 while intact glycopeptides can be analyzed by MS with or without enrichment,129 although the enrichment step is beneficial for deep characterization of glycopeptides with low stoichiometry.130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145 Historically, chemical enrichment methods were adopted for both glycans and glycopeptides such as hydrazide chemistry,146,147,148,149 boronic acid,142,150,151,152 etc. Hydrophilic interaction liquid chromatography (HILIC),100,153,154,155 lectin affinity chromatography,121,156,157 and graphitized carbon chromatography are the most widely adopted methods for enrichment of glycopeptides.124,158,159,160,161
LC-MS/MS analysis (Fig. 2, middle). Before tandem MS/MS analysis, chromatographic separation can simplify the composition of glycans and glycopeptides.162 The underivatized native glycans are hydrophilic and usually separated by PGC columns,163,164,165 while the permethylated glycans are hydrophobic and often separated by reversed-phase C18 chromatography.166,167 For the separation of glycosite-containing peptides and intact glycopeptides, the reversed-phase C18 chromatography,166,168,169,170,171,172,173,174,175 HILIC,176,177,178 and PGC are also widely used.179,180 Moreover, cation-exchange chromatography (CEX),181,182 size-exclusion chromatography (SEC),183,184,185 and capillary electrophoresis (CE) are also applied to the separation step.186,187,188
The separated glycans or glycopeptides are then analyzed by tandem MS/MS with various dissociation methods97,189,190,191,192 (Fig. 2, middle). Because of the possible appearance of multiple putative glycosites on a single peptide and the frequent presence of structural isomers in glycans,165,193 glycosite localization and glycan structure identification are the two major challenges in MS/MS analysis of both N- and O- glycosylation. A general workflow for the MS/MS analysis of intact glycopeptides are shown in Fig. 3 (note that this workflow is a stereotyped summarize of schemes for the identification of intact glycopeptides, which means many studies will not strictly follow this workflow). The N-glycosites can be localized by site-determining fragment ions from MS2 spectra,194 and structural isomers are distinguished with structure-diagnostic fragment ions of the N-glycan moieties.195 While O-glycosites, due to the frequent existence of three target amino acids (S/T/Y) and densely glycosylated adjacent sites, are much more difficult to be determined than N-glycosites.196,197,198,199,200,201,202,203
N-glycosites rigorously follow the motif rule of N-X-S/T/C (X represents any amino acid except proline),204,205 and several other peculiar motif rules have been reported.99,206,207,208 Therefore, the methods for acquiring evident MS2 signals and the algorithms of parsing the MS2 data are necessary.209,210,211,212,213
A precise site-determination method for N-glycosites is to use paired b*/y* ions in the MS2 spectrum,214 where b* and y* ions respectively refer to peptide b and y ions with a connected GlcNAc residue215 (Fig. 3b). These b* and y* ions often appear with moderate abundance after the cleavage of glycopeptide precursor ions.216 Several detected b* or y* ions can narrow the possible area of N-glycan moiety and make it covers only one putative site so that the real N-glycosite can be determined.72,217,218
Collision-based dissociation, such as collision induced dissociation (CID) and higher energy collisional dissociation (HCD),219 can cause peptide fragmentation (either glycosite-containing peptides or intact glycopeptides) and produce abundant b/y fragment ions. Electron-based dissociation, such as electron capture dissociation (ECD)220 and electron transfer dissociation (ETD),221 has the advantage of causing “gentle” dissociation of the peptide backbone without neutral loss of the N-glycan moiety, generating c/z ions. Ultraviolet photodissociation (UVPD),222 simultaneously including the features of both collision- and electron-based dissociation, provides comprehensive types of fragment ions. Selective fragmentation of either the peptide backbone or the N-glycan moiety can be achieved with the combination of different dissociation methods (such as ETD + CID) or different energies of the same dissociation method (such as high and low normalized collision energies of HCD).223,224,225,226,227 In addition, combinatory dissociation methods such as combined EThcD and ETciD on Orbitrap mass spectrometers have also been applied.228,229
N-glycans on N-glycoproteins contain hundreds of compositions and more than ten thousand different structures in mammals.230 N-glycosylation occurring on an identical site of glycoprotein may have thoroughly different biological processes because of distinct monosaccharide compositions.231,232,233,234,235,236,237,238,239,240,241,242 Even N-glycans sharing the same monosaccharide composition may have different functions due to the glycan structures,243,244 indicating that the significant roles of N-glycan structures in regulating the functions of N-glycoproteins.245 Therefore, structure-specific characterization of N-glycans is urgently needed at both aspects of chemistry and biology.246 In general, tandem MS/MS analysis of intact N-glycopeptides is able to precisely identify peptide backbone sequences, N-glycosites as well as N-glycan compositions and structures.247,248 However, due to the limitation of MS analytical discernibility, some monosaccharide isomers are unable to be distinguished. For example mannose, galactose, and glucose are interpreted as hexoses in glycan compositions.249 Moreover, N-glycans with the same monosaccharide composition may as well form different structures with different amount of antenna and serial numbers of linked carbon atoms (β-1,2 or β-1,4 at α-1,3 core mannose, etc.).87,250,251
To unambiguously discriminate the structural isomers, a pivotal series of fragment ions in MS2, herein named structure-diagnostic ions are required.252 This kind of ions are in fact the fragmented N-glycan A/B/C/X/Y/Z ions which can independently distinguish a specific structure from the structural isomers.253,254 N-glycan structures can be discriminate by detecting theoretical structure-diagnostic ions which are generated in silico relying on the theoretical N-glycan structure database created by the retrosynthetic strategy,78 and structures of intact N-glycopeptides are figured out by assigning N-glycan structures to peptide backbones.255
Bioinformatics analysis (Fig. 2, right). For identifying intact glycopeptides from LC-MS data, two strategies of MS data acquisition have currently been adopted: data-dependent acquisition (DDA) and data-independent acquisition (DIA).256,257,258 Most of the software and platforms for analyzing intact glycopeptides are designed to search against the spectra generated from DDA.259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280 DDA focuses on the precursors with high intensity and specifically isolates them to form fragments and generate MS2 spectra.281,282,283 Based on the DDA data, the method for identifying intact N- and O-glycopeptides, which consists of the following steps (the order of these steps may be rearranged in several platforms or software): (1) deducing peptide backbone by peptide fragment ions, (2) determining glycan mass by calculating the mass difference between deduced peptide backbone and intact glycopeptide precursor, (3) localizing glycosite by matching specific glycosite-containing ions, and (4) characterizing glycan composition or structure using glycan or glycan-containing fragment ions, is adopted by most of the software such as Byonic,284 pGlyco,285,286 GPQuest,287 GPSeeker,252 O-pair Search in MetaMorpheus,288 MSFragger-Glyco289, and StrucGP.248 In-silico digested theoretical peptide database or customized experimental peptide spectra library is used for the identification of peptide backbone of intact glycopeptide.290
The characterization of glycans (especially N-glycans) by DDA can be achieved by several strategies (Fig. 3a), including (1) parsing N-glycan compositions using theoretical glycan composition database (for instance Byonic,284,291,292,293,294 which calculates the precise masses of glycans constructed by proper combinations of monosaccharides, giving the number of Hex, HexNAc, etc). These compositions together with their masses are then stored in the composition database and the exact masses of relevant theoretical fragment ions are also calculated and matched in the MS2 spectra for further characterization; (2) parsing N-glycan structures using theoretical structure database built by retrosynthesis rules (for instance GPSeeker72), and (3) parsing N-glycan structures using de novo algorithm (for instance StrucGP248). The first strategy only offers the information of monosaccharide composition, while the second and third strategies can provide N-glycan structure information. In particular, the second strategy uses structure-diagnostic ions to distinguish different theoretical structures from the same monosaccharide composition and provides the entire structure of each characterized N-glycan. In contrast, the third strategy sequentially matches a series of Y ions and complementary B ions to form an intact N-glycan structure (that is, de novo algorithm), and shows structures with high accuracy regardless of theoretical database. However, the third strategy may ambiguously distinguish symmetrical structures in some applications. StrucGP is the first search engine that adopts de novo algorithm to conduct structure-specific identification of intact glycopeptides.248
The application of DIA to identify intact glycopeptides is still very young.295,296 Compared with the DDA strategy, DIA does not select specific precursors based on MS1 peak intensities.256 Instead, DIA collects all ions acquired in MS1 based on retention time and fragments these ions to generate MS2 spectra,256,295 suggesting that LC-MS/MS data from DIA contains complete information of the sample rather than DDA data which only contains the information of peptides with high abundance. However, interpreting DIA data remains a challenge and needs more advanced algorithms such as machine learning.297,298 The techniques adopted to analyzing DIA data includes pre-building corresponding DDA data library and many other methods.296,299,300 As for the identification of intact glycopeptide using DIA strategy, SWATH-MS workflow has also been adopted,301 and the characterization of glycosylation has been achieved at the molecular levels of intact glycopeptide and glycan.302,303
Glycosylation of SARS-CoV-2 proteins
The aforementioned high-throughput detection and analysis of the structure and localization of protein glycans is a prerequisite for discovering and studying the function of glycosylation,28,87,304 which will lead to a better understanding of glycoprotein functions and the molecular mechanisms of infectious disease.38,50,305,306,307
SARS-CoV-2 is a positive-sense single-stranded RNA virus.308,309 Sequence analysis of SARS-CoV-2 isolates shows that the 30 kb genome at least encodes 29 proteins, including 4 structural proteins, 16 non-structural proteins (NSP1-NSP16), and 9 accessory factors (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c, ORF10).310,311 The NSPs involve in virus processing and replication,312,313,314,315 while the structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N) are responsible for host recognition, binding, recycling, and pathogenesis.34,36,37,38,39,40,41,42,316 According to the in silico topology, the majority of the encoded proteins are glycoproteins, although only four of them have been reported with their exact glycosites to date.
S protein
Among the structural proteins, S protein in SARS-CoV-2 is the only one with sequence variability >20% when compared with SARS-CoV.317 It is a trimeric transmembrane protein that composes of two functional subunits S1 and S2.318,319 The S1 subunit is responsible for host cell receptor binding, while S2 subunit is for membrane fusion.320,321 The total length of S protein is 1273 amino acids, and the receptor binding domain (RBD) is located in the region from amino acid 319 to 541 in S1 subunit.322 The receptor binding motif (RBM) that mediates the contact with the angiotensin-converting enzyme 2 (ACE2) receptor locates in the RBD from amino acid 437 to 507.320,321,323 The S protein can recognize and bind to ACE2 receptor as the primary host cell infection route.324 Therefore, S protein determines the infectivity and transmissibility of SARS-CoV-2 and is the major antigen and target of vaccination.325,326
S protein is a well-known glycoprotein, and the modified glycans shield about 40% of the protein surface of the S trimer,35 which functions as camouflage to humoral and cellular components of the host innate immune system.54 Compared with Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV, the S protein of SARS-CoV-2 has a lower glycosylation density,35,63,327 indicating the S protein surface is more exposed and it is more effective in eliciting humoral immunity.31 Since the first report of 16 N-linked glycosites by cryo-electron microscopy (cryo-EM),320 the characterization of glycosylation of S protein becomes a hotspot.38,62,63,70,145,328,329,330,331,332,333,334,335,336 In total, 23 N-linked glycosites with high occupancy (mostly >95%) have been reported (Fig. 4).40,62,63,70,328,329,330,331,332,333,334,335 In contrast, among all the O-linked glycosites, only two sites show relative high occupancy (Table 1).62,63,70,80,328,332,333,336,337,338,339,340,341,342 The S1 subunit has 13 putative N-glycosites (N17, N61, N74, N122, N149, N165, N234, N282, N331, N343, N603, N616, and N657) with the N-X-S/T (X ≠ P) sequon, one putative N-glycosite (N334) with the N-X-C (X ≠ P) sequon and two putative O-glycosites (T323 and S325), of which T323, S325, N331, N334, and N343 are located on RBD. The S2 subunit has 9 putative N-glycosites (N709, N717, N801, N1074, N1098, N1134, N1158, N1173, and N1194) with the N-X-S/T (X ≠ P) sequon.
Site-specific N-glycan types of recombinant SARS-CoV-2 S proteins expressed in human cells (a), insect cells (b), or from native S protein (c). The Y-axis of the histogram refers to the number of published papers that report the corresponding sites with detailed site-specific N-glycan types. The proportion in the pies represents the glycan types reported in the relevant papers. The region of RBD and the furin cleavage site are marked
Although the N-glycosites of S protein identified by different teams in different expressed systems are almost same, the glycan compositions and structures as well as their occupancy are distinct (Fig. 4). MS-based characterization of recombinant S protein expressed in human cells including human embryonic kidney (HEK) 293F cells and HEK 293 cells shows that the glycans on N234 and N709 are mainly oligomannose-type.32,40,63,70,329 Complex-type glycans can be predominantly found at N17, N74, N149, N165, N282, N331, N343, N616, N657, N1098, N1134, N1158, N1173, and N1194 residues, while six positions including N61, N122, N603, N717, N801, and N1074 are modified by a mixture of oligomannose- and complex-type glycans.63 Notably, the most common oligomannose-type glycan is Man5GlcNAc2. More than half of these N-linked glycans are fucosylated,63 and highly processed sialylated complex-type glycans can be predominantly found on the residues of N165, N282, N801, N1074, and N109870,332 (Fig. 5). By using energy-optimized LC–MS/MS method, glycoforms including the LacdiNAc and polyLacNAc structural motifs have been revealed on N-glycans of S protein expressed in the HEK293 expression system.225,332 Moreover, a recent quantitative N-glycan analysis on protein of S1 subunit purified from SARS-CoV-2 infected Calu-3 cells by immunoaffinity purification showed that the complex-type N-glycans (79%) with 21% oligomannose and/or hybrid structures predominate.343 In addition to the diverse N-linked glycans of S protein identified by MS, the N-linked glycan structures of RBD of S glycoprotein expressed in human HEK293F cells have been characterized by nuclear magnetic resonance (NMR) spectroscopy, which avoids sample digestion and derivatization.330 A lot of glycan structures missed in MS-based approaches have been disclosed.330 Besides the expected N-acetyllactosamine (LacNAc), 3′SLN (3′SLacNAc), and 6′SLN (6′SLacNAc) terminal moieties at the glycans of N331 and N343, the unprecedented structures such as LeX (LewisX), LDNF (LeX and fucosylated lacdiNAc), and 6′SLDN (6’SLacdiNAc) were also identified.330
Compiled structures of N-glycans identified on 23 N-glycosites of S protein. Studies were summarized only when the reported N-glycans were characterized by fragment ions in MS2 spectra that can help deduce their structures or N-glycans were exhibited with annotations to reveal their exact structures. A little ambiguousness in N-glycan structures such as uncertain position of terminal monosaccharide are allowed. Studies that reported N-glycans with only monosaccharide compositions were not placed in this figure
The distinct types of N-linked glycans of S protein in different expression systems are proposed to be determined by the differential processing of N-glycans among different species, rather than the location of glycosites.62,342 Different from that in human cell expression system (Fig. 4a),40,62,63,70,328,329,330,332 the N-linked glycans of recombinant S protein obtained from insect cell expression system are most high mannose-type62,329,333,334 (Fig. 4b). It is worth noting that besides the N331 and N343 in the RBD, glycosylation at N334 has also been detected in recombinant S protein expressed from Spodoptera frugiperda (Sf9) cells with low occupancy,333 which is consistent with the report of an N-X-C motif exhibiting substantial N-glycosylation344 (Fig. 4b). Interestingly, compared with the recombinant S protein in insect cells, native S protein has lower levels of oligomannose-type glycans and high levels of complex-type glycans331 (Fig. 4c). These studies collectively generated a comprehensive N-glycosylation map of the S protein, and all the identified structures are plotted in Fig. 5.35,40,62,63,70,145,328,329,330,331,332,333,335,336,341,342,343,345
The N-linked glycosites of S protein have diverse functions (Fig. 6). N165 and N234 are located near the RBD.63,346 N-linked glycosylation at N234 is largely accessible to α-1,2-mannosidases and can regulate the conformational dynamics of RBD.34 Deletion of the glycans through N165A and N234A mutations significantly changed the RBD conformational shift towards to the “down” state (presenting a receptor-inaccessible state) and reduced the binding to ACE2, which suggests that the glycosylation at N165 and N234 may promote host recognition.34 Moreover, N282, N331, and N343 are also proximal glycosites that shield the receptor binding sites of S protein, especially the RBD in the “down” state.63,70,320,347 Besides the involvement of these glycosites in the binding of SARS-CoV-2 to the receptor, these glycosites also affect the sensitivity of viruses to neutralizing antibodies. For example, N234Q mutation can significantly decrease the sensitivity to neutralizing antibodies, whereas N165Q and N709Q mutations increase the sensitivity.2 In addition, N149Q, N331Q, and N1173Q mutations also dramatically increase the sensitivity to convalescent sera,39,316,348 indicating the influence of glycans in epitopes targeted by neutralizing antibodies (Fig. 6). The glycosylation is also important for virus virulence and viral infection. When the glycosylation at N122, N331 with N343, N717, N801, and N1074 of S protein are inhibited by mutations, the viral infectivity of SARS-CoV-2 is significantly reduced39,316 (Fig. 6). The polybasic cleavage site (RRAR) at the junction of S1 and S2 subunits is one notable feature of SARS-CoV-2, which is not observed in SARS-CoV.349,350 The RRAR can be cleaved by furin or other proteases and play important roles in determining viral fusion, entry and pathogenesis.36,351,352 The N-glycosylation at N61, N603 and N657 is proximal to the furin-site and able to increase the steric hindrance for cleavage, which seems to be beneficial to SARS-CoV-2 entry36 (Fig. 6). Different from being cleaved by proteases, computational saturation mutagenesis of N616 and N1134 residues increases the stability of S protein,353 which may be associated with glycosylation; however this phenotype needs experimental confirmation (Fig. 6). 3D structural modeling of glycosylated SARS-CoV-2 trimmer S protein disclosed the micro-heterogeneity of N-glycosites. The glycans at N74 and N165 residues of S protein interact with ACE2 receptor glycan at N546 residue and thus modulate Spike-ACE2 interactions, suggesting that the changes of glycans occupancy may affect the affinity and alter the infectivity40 (Fig. 6).
Glycosites of S protein and their functions. a The functions of glycosylation of S protein. Different shapes and colors represent the corresponding functions, and the gray indicates that the functions of the sites are unknown. b Structure-based display of the N-glycosites in the S protein. The glycosites of S protein are marked on the three-dimension structures. S protein is shown in the “RBD up” state. A top view and a side view (up) of the S protein (PDB: 6VYB) are presented. N74 and N165 are not labeled due to the information missed in the crystal structure331
Besides the N-glycosites, a large number of putative O-glycosites have also been found in S protein (Table 1). Among them, the levels of O-glycosylation at T323 and S325 are relative higher, while the glycosylation of other O-glycosites are in low occupancy.40,63,70,329,332,336,337,338 The O-glycans identified on O-glycosites of S protein and RBD from human cells as well as deduced O-glycan structures were summarized70,80,328,332,336,338,340,341,342,354,355,356,357 (Table 2, Table 3). O-linked glycans such as Core-1,336 disialylated Core-1,332 Core-2,328 mucin-type,339 and sialylated mucin type are reported on the recombinant S protein.70
Although various O-glycosites have been identified, their functions remain largely unknown. Similar to N-glycosylation, the functions of O-glycosylation of S protein are also very important. More than 60% O-glycosites located close to N-glycosites may suggest the possible complementary functions of O-glycans in immune shielding.336 The furin cleavage site is unique to the SARS-CoV-2 S protein compared to SARS-CoV. The O-glycosylation at T678, adjacent to the polybasic furin cleavage site, carries Core-1 and Core-2 structures capped primarily with α2–3 sialic acid, which may suggest that cleavage is potentially regulated by the nearby O-glycans.332,358 Mutation of N616 abolished the O-glycosylation at T618 indicates that N-glycosylation at N616 is the prerequisite of N-sequon-related O-glycosylation, which obeys an “O-Follow-N rule”.80 T323 and S325 residues are two conserved O-glycosites in the RBD of S1 subunit, which may play important roles in mediating Spike-ACE2 binding.62,70,328 Compared with SARS-CoV, S494 is one of the six mutations on the RBD of S protein encoded by SARS-CoV-2. Attachment of the O-glycans to S494 can increase the binding affinity of virus to ACE2.354 The predicted O-linked glycosylation residues at S673, T678 and S686 are near the RRAR position,36,337 implying their potential functions in virus penetration.332,337,359
E protein
E protein is a small integral membrane protein of 8–12 kDa in SARS-CoV-2,360 and functions in viral assembly, release and pathogenesis.361,362,363 It comprises of three domains, including a short hydrophilic N-terminus domain, a hydrophobic transmembrane domain and long hydrophilic C terminal region.364,365 Previous studies showed that E proteins in many coronaviruses can form pentameric structures exhibiting cation selective channel activity,366,367,368,369 which is critical for viral infectivity370,371 and Ca2+ conductivity in the ER-Golgi intermediate compartment.372 Based on the sequence prediction, two putative N-linked glycosites may exist in the transmembrane segment of E protein at positions N48 and N66373 (Fig. 7a). Probably due to the proximity of the residue to the membrane, residue N48 is difficult to be glycosylated.361 In contrast, N66 is found to be modified with oligomannose-type glycans.373 Mutation of residue N66 can promote the resembling of dimers and trimers of E protein which is required for virion assembly, while the monomer may function in disruption of the host secretory pathway.361
M protein
M protein is the most abundant envelope protein of SARS-CoV-2 that contains 222 amino acids.374,375 It comprises of three N-terminal transmembrane domains,374 and is essential for the assembly of virus particles by interacting with other three structural proteins of SARS-CoV-2.376,377,378 Similar to E protein, the glycosylation of M protein has not yet been extensively studied and characterized. In silico computation and simulation has revealed the topology of M proteins from different coronaviruses and predicted eight N-glycosites including N5, N21, N41, N43, N117, N121, N203, and N216 were predicted379 (Fig. 7b). The functions of these N-linked glycosylation remain to be studied.
ORF3a protein
The non-structural proteins of human coronaviruses are indispensable for viral replication and transcription.312,380 ORF3a is a non-structural protein of SARS-CoV-2 localized at the surface. It is the largest accessory factor that contains 274 amino acids,381 and shows broad functions,382,383 such as enhancing viral entry within the host,381 regulating the pro-inflammatory cytokine and chemokine production,384 participating in ion channel formation as well as modulating release of virus from the host cell.381 According to the hydrophobicity analyses and topology studies, there may be four O-linked glycosites at S27, T28, T32 and T34 residues,385,386 with higher O-glycosylation occupancy at T28 and T32 residues; N-glycosylation is absent in ORF3a protein382 (Fig. 7c). The functions of these O-linked glycosylation remain to be investigated.
Glycosylation of human target protein ACE2
The severity of SARS-CoV-2 infection varies greatly among individuals.387 One possible reason may be due to the different expression of SARS-CoV-2 receptor.40 ACE2 is the main human receptor of SARS-CoV-2.388,389 It is expressed on the membrane of cells located in many organs (such as heart, kidney, and intestines) and is a promising drug target.390,391 Besides expression difference, glycosylation on ACE2 also affects the SARS-CoV-2 entry and infectivity.392 In the recombinant ACE2 protein from HEK293 cells, 7 N-glycosites and 2 O-glycosites have been identified (Fig. 8). The majority of glycans at N53, N90, N103, N322, N432, N546, and N690 residues of ACE2 are of complex-type, always with >75% occupancy, and the sialic acid linkage always exist in the glycans.36,40,345 The sialic acid was previously identified to serve as an attachment factor for a number of coronaviruses including MERS-CoV,393 transmissible gastroenteritis virus,394 human coronavirus (HCoV)-OC43,395 and HCoV-HKU1.396,397 The sialic acids present on ACE2 substantially block infection of SARS-CoV; however, the block effect is much smaller in the case of SARS-CoV-2.398 In particular, N-glycosylation at residues N90, N322, and N546 of ACE2 play critical roles in the binding of ACE2 with RBD of S protein. Mutation of N90 residue increases the binding affinity to S protein,399 indicating N90 glycosylation can protect host cells against viral infection.400 Atomistic molecular dynamics (MD) simulations show that N322 glycan binds to the core region of RBD of S protein from amino acid 369 to 378.40,400 The interaction between RBD and N322 residue of ACE2 is much stronger than that between RBD and N90 residue of ACE2. Besides, the antibody (CR3022) obtained from SARS-CoV infected patients has a binding site that overlaps remarkably with that of the N322 glycan, suggesting the N322 glycosylation may affect viral infection.400 Moreover, MD simulations show that N546 residue involves in the glycan-glycan interactions with S protein at N74 and N165.40 Of the two identified O-glycosites of ACE2, the stoichiometry of glycosylation at S155 is extremely low, and the function remains elusive.40 T730 residue is distal to the binding interface between S protein and ACE2, and the Core-1 mucin type O-glycan GalNAcGalNeuAc2 is the predominant glycan on it. It is speculated that the massive hydrophilic glycosylation at T730 in the juxtamembrane region outside the cell membrane may affect the dimerization and the presentation of ACE2 on the cell surface.345
Therapeutic strategies for COVID-19 associated with glycosylation
As of 5 April 2021, there are 216 vaccines and 506 therapeutic drugs at different development stages for COVID-19. Among them, 92 vaccines and 419 therapeutic drugs are undergoing clinical trials, and 122 vaccines and 87 therapeutic drugs are in preclinical development (https://biorender.com/covid-vaccine-tracker). Influence of site- and structure-specific glycosylation on infectivity and immune escape is one of the key factors for vaccine development.31,41,306 The efficacy of some vaccines and therapeutic drugs may be closely associated with glycosylation (Table 4).
Neutralizing antibodies
The neutralizing antibodies are one the most important specific defense against viral infection.348,401,402 Antibodies that specifically target viral proteins can block the interaction between the virus and the host cell, thereby preventing the virus entry for replication.403,404 By high-throughput single-cell sequencing of COVID-19 patients’ B cells, potential SARS-CoV-2 neutralizing antibodies have been found from convalescent patients such BD23-Fab.348 Glycosylation at the N165 of S protein can facilitate the binding of BD23-Fab to the RBD.348 S protein has highly conserved glycosylation patterns between SARS-CoV and SARS-CoV-2, the antibodies bound to glycopeptide epitopes of SARS-CoV are critical for the screening of monoclonal antibody (Mab) to treat SARS-CoV-2, such as MAb S309 that has been isolated from SARS-CoV patient targeting an epitope containing a glycan at N343.334,405,406 Notably, the antibodies isolated from patients recovering from SARS-CoV, such as the monoclonal antibodies VIR-7831 (Phase II clinical trial), VIR-7832 (Phase I/II clinical trial) and their parent antibody (S309), can also effectively neutralize SARS-CoV-2 in vivo and in vitro.406,407,408 Besides the antibodies from the recovered patients, natural antibodies formed spontaneously without specific immunization may also be very useful for SARS-CoV-2 treatment.409,410 GalNAc-O-Ser/Thr (Tn antigen) and Gal-GalNAc-O-Ser/Thr (T antigen) are well-known natural antigens and associated with the pathogenesis of many diseases.411,412,413 Compared to non-infected individuals, the anti-Tn antibodies level in COVID-19 patients are significantly lower, suggesting that natural anti-Tn antibodies may be protective against COVID-19.414 In addition, the HIV-1 Env Fab-dimerized glycan (FDG)-reactive antibodies are an anti-glycan antibody that recognize high mannose glycans of SARS-CoV-2, indicating the potential prospects of these natural antibodies in SARS-CoV-2 treatment.415
Vaccines
Vaccination is the most effective long-term strategy for the prevention and control of COVID-19.6,416 Vaccines, such as inactivated vaccines,417,418,419 DNA plasmid vaccines,420,421 adenovirus-vectored vaccines,422,423 RNA vaccines,424,425 protein subunit vaccines,333,426 and virus-like particle vaccines,427,428 have been developed. In the protein subunit vaccines, the RBD of S glycoprotein is an ideal immunogen.333,429,430 Because of the existence of glycosites in the immunogenic epitope of the virus, the immunogenic epitopes masked by glycosylation may not be recognized by the host, thus leading to immune escape of the virus.431 By mapping the glycosites on the complex structure of the RBD bound to ACE2, it is found that most glycosites are located in the RBD core subdomain and distant from the bound ACE2, indicating that glycans on RBD may not affect receptor recognition and/or binding.333 In addition, the viral glycans are also important immunogens.432,433 The complex N-glycans such as GlcNAc2-4-Man3-GlcNAc2-Asn in N74, N149, N282, N801, N1074, and N1098 of S protein,70,434,435 as well as oligomannose-type glycan Man5-GlcNAc2 in N234 may be suitable immunogens for developing vaccines.40,70,329,434,435
Other drugs
Lectins are carbohydrate-binding proteins binding to sugar groups, and have potent antiviral properties through preventing the attachment of virus to host cell.59,436,437 FRIL is a lectin isolated from hyacinth beans and serves as an antiviral agent by blocking the complex-type N-glycans against SARS-CoV-2.438 Griffithsin, a red-alga-derived lectin, is in phase I clinical trial for the treatment HIV infection and also is promising for the treatment of COVID-19 by binding to the oligosaccharides on the surface of viral glycoproteins.439,440,441 Other lectins such as Clec4g and CD209c can also bind to the N-glycans of S protein and interfere the Spike-ACE2 interaction and reduce SARS-CoV-2 infection.442 Notably, Lentil lectin derived from Lens culinaris can bind specifically to oligomannose-type glycans and GlcNAc at the non-reducing end terminus of S protein, thus block the binding of ACE2 to S trimer, showing the strongly inhibit infection of SARS-COV-2, including epidemic variants B.1.1.7, B.1.351, and P.1.443
The major cause of death by SARS-CoV-2 refers to the “cytokine storm”,384,444,445,446 which is featured as excess release of inflammatory cytokines, such as interleukin (IL)-1, tumor necrosis factor α (TNF-α), and IL-6.447 Galectin-3 (Gal-3), a member of β-galactoside-binding lectins that preferentially binds to N-acetyllactosamine moieties on glycoconjugates, showed a dramatic increase with cytokine storm.448,449 Inhibition of Gal-3 can reduce the releases of IL-1, TNF-α, and IL-6 from macrophages, suggesting Gal-3 inhibitor as a promising agent for COVID-19 treatment.450,451 Currently, the Gal-3 inhibitor TD139 is undergoing clinical trials for the treatment of COVID-19,452,453 and other Gal-3 inhibitor such as GB1107,450,454 belapectin (also called GR-MD-02) are under investigation.455
Iminosugars, also called iminosaccharides, are the analogs of common sugars where an oxygen atom is replaced by a nitrogen atom in the ring of the structure.456 They are known to interfere with the N-linked glycosylation by inhibiting the α-glucosidase I and II enzymes on the ER,457,458 thus affecting the interaction between viral glycoproteins and host receptor. Iminosugars such as Celgosivir, Castanospermine and the monocyclic UV-4 have been reported to prevent SARS-CoV-2-induced cell death and reduce viral replication,459 while Miglustat can lead to a dramatically decrease of the viral Spike protein of SARS-CoV-2.460 Other potential inhibitors with similar structures such as α-mannosidase inhibitors Kifunensine also show similar roles in reducing SARS-CoV-2 entry.36,461
DAS181 is a kind of inhaled bacterial sialidase that functions by removing sialic acid from the surface of epithelial cells, thus preventing attachment and subsequent infection by respiratory viruses.462,463 The sialic acid linkage always existed in the glycans of ACE2,40,345,392 suggesting the potential therapeutic effect of DAS181 in COVID-19 treatment. Currently, DAS181 is in phase III clinic trial for patients with severe COVID-19.
Perspectives
It is well-known that virus may alter the glycan coat on the viral surface to enhance the infectivity and affect immune recognition.50,464 With the rapid development of techniques for characterizing the glycans and the glycoproteins,219,289 the biological functions and significance of glycans and glycoproteins of virus are disclosed, which broads the understanding of virus biology.38,45,464 As described above, both SARS-CoV-2 proteins (especially S and N proteins) and their receptor (ACE2) are densely glycosylated. The glycan masses on S protein, N protein and ACE2 are about 80 kDa,333 13 kDa,465 and 30 kDa,466 respectively; the average mass of a single glycan is about 4 kDa, indicating that these proteins are glycosylated simultaneously on multiple sites, although other modifications, such as phosphorylation,465 may also contribute to the extra masses on the basis of the protein sequence. Characterization of glycosylation at the intact N-glycopeptide level with the assistance of state-of-the-art enrichment will deliver comprehensive glycosylation information (glycosite, glycan composition and structure) for single sites,289 the cross-talk between different glycosites as well as other PTMs previously missed.80,467 Adoption of protein enzymes (such as Glu-C, Asp-N other than trypsin or chymotrypsin) cutting less frequently occurring amino acids to produce larger and longer peptides,468 or no enzyme at all (i.e, the top-down method) may be an optional choice.469 However, delicate selective dissociation of peptide backbones and glycan moieties as well as versatile bioinformatics tools supporting interpretation of multiple modifications at a time needs to be developed in the future.
The evolution of SARS-CoV-2 is fast within the human population by gaining fitness-enhancing mutations, which may alter viral infectivity and disease severity, and escape the host immunity even in individuals who have been vaccinated. For example, mutation of D to G at the residue 614 (D614G) of S protein moderately increases the infectivity and transmissibility.331,470,471,472 Following the D614G mutation, N439K and Y453F mutations within the RBM of S protein appears in SARS-CoV-2 variants. These mutations not only enhance the binding affinity for the ACE2 receptor, but also reduce the therapeutic efficacy of neutralizing antibodies.473,474 SARS-CoV-2 Delta variant, also known as lineage B.1.617.2, is a variant of lineage B.1.617 of SARS-CoV-2. It has three mutations on S protein including T478K, P681R and L452R, which dramatically increases transmission and leads to antibody escape.475,476,477 However, despite many SARS-CoV-2 variants appear, whether the mutations of SARS-CoV-2 variants would affect the glycosylation profile of SARS-CoV-2 is still less understood. Given the critical roles of glycosylation in host recognition, penetration, binding, recycling and pathogenesis, uncovering the glycosylome of SARS-CoV-2 variants may help to increase the understanding of viral biology and develop more effective vaccines and drugs for SARS-CoV-2 variants.
Data availability
No additional data are included.
References
Wang, D. et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. Jama 323, 1061–1069 (2020).
Li, Q. et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182, 1284–1294.e1289 (2020).
Tay, M. Z. et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 (2020).
Udugama, B. et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano 14, 3822–3835 (2020).
Rajendran, D. K. et al. Systematic literature review on novel corona virus SARS-CoV-2: a threat to human era. Virusdisease 31, 161–173 (2020).
Hu, B., Guo, H., Zhou, P. & Shi, Z. L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154 (2021).
Rabaan, A. A. et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez. Med. 28, 174–184 (2020).
Petrosillo, N. et al. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 26, 729–734 (2020).
Petersen, E. et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 20, e238–e244 (2020).
Gates, B. Responding to Covid-19 - a once-in-a-century pandemic? New Engl. J. Med. 382, 1677–1679 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Yang, X. et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 8, 475–481 (2020).
Lv, M. et al. Coronavirus disease (COVID-19): a scoping review. Euro Surveill. 25, 2000125 (2020).
Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA 323, 1239–1242 (2020).
Xu, X. et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imaging 47, 1275–1280 (2020).
Li, L. Q. et al. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J. Med. Virol. 92, 577–583 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
V’Kovski, P. et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021).
Amanat, F. & Krammer, F. SARS-CoV-2 vaccines: status report. Immunity 52, 583–589 (2020).
Jin, Y. et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 12, 372 (2020).
Mari, A. et al. Reply to Nikolaos Grivas, Sanchia Goonewardene, Wouter Everaerts, Nikolaos Kalampokis’s Letter to the Editor re: Andrea Mari, Riccardo Tellini, Francesco Porpiglia, et al. Perioperative and mid-term oncological and functional outcomes after partial nephrectomy for complex (PADUA Score ≥10) renal tumors: a prospective multicenter observational study (the RECORD2, Project). Eur Urol Focus. In press. https://doi.org/10.1016/j.euf.2020.07.004. Eur Urol Focus S2405-4569, 30306 (2020).
Shang, J. et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA 117, 11727–11734 (2020).
Parasher, A. COVID-19: current understanding of its pathophysiology, clinical presentation and treatment. Postgrad. Med. J. 97, 312–320 (2021).
Wenjie, L. et al. The mechanism of glycosylation in SARS-CoV-2 infection and application in drug development. Prog. Chem. 33, 524 (2020).
Spiro, R. G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43r–56r (2002).
Spiro, R. G. Glycoproteins. Adv. Protein Chem. 27, 349–467 (1973).
Hart, G. W. Glycosylation. Curr. Opin. Cell Biol. 4, 1017–1023 (1992).
Pinho, S. S. & Reis, C. A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015).
Reily, C., Stewart, T. J., Renfrow, M. B. & Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019).
Shental-Bechor, D. & Levy, Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc. Natl Acad. Sci. USA 105, 8256–8261 (2008).
Zhao, X., Chen, H. & Wang, H. Glycans of SARS-CoV-2 spike protein in virus infection and antibody production. Front Mol. Biosci. 8, 629873 (2021).
Chatterjee, S. K. & Saha, S. Biotechnology to Combat COVID-19 (IntechOpen, 2021).
Shajahan, A. et al. Glycosylation of SARS-CoV-2: structural and functional insights. Anal. Bioanal. Chem. 1–15, https://doi.org/10.1007/s00216-021-03499-x (2021).
Casalino, L. et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 6, 1722–1734 (2020).
Grant, O. C., Montgomery, D., Ito, K. & Woods, R. J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 10, 14991 (2020).
Yang, Q. et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife. 9, e61552 (2020).
Jennings, B. C., Kornfeld, S. & Doray, B. A weak COPI binding motif in the cytoplasmic tail of SARS-CoV-2 spike glycoprotein is necessary for its cleavage, glycosylation, and localization. FEBS Lett. 595, 1758–1767 (2021).
Ramírez Hernández, E. et al. The role of the SARS-CoV-2 S-protein glycosylation in the interaction of SARS-CoV-2/ACE2 and immunological responses. Viral Immunol. 34, 165–173 (2021).
Wang, L., Wang, L. & Zhuang, H. Profiling and characterization of SARS-CoV-2 mutants’ infectivity and antigenicity. Signal Transduct. Target Ther. 5, 185 (2020).
Zhao, P. et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28, 586–601.e586 (2020).
Reis, C. A., Tauber, R. & Blanchard, V. Glycosylation is a key in SARS-CoV-2 infection. J. Mol. Med. 99, 1023–1031 (2021).
Nunes-Santos, C. J., Kuehn, H. S. & Rosenzweig, S. D. N-glycan modification in Covid-19 pathophysiology: in vitro structural changes with limited functional effects. J. Clin. Immunol. 41, 335–344 (2021).
Schjoldager, K. T., Narimatsu, Y., Joshi, H. J. & Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 21, 729–749 (2020).
Mariño, K., Bones, J., Kattla, J. J. & Rudd, P. M. A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 6, 713–723 (2010).
Vigerust, D. J. & Shepherd, V. L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15, 211–218 (2007).
Sugrue, R. J. Viruses and glycosylation: an overview. Methods Mol. Biol. 379, 1–13 (2007).
Carbaugh, D. L. & Lazear, H. M. Flavivirus envelope protein glycosylation: impacts on viral infection and pathogenesis. J. Virol. 94, e00104–20 (2020).
Tannous, A., Pisoni, G. B., Hebert, D. N. & Molinari, M. N-linked sugar-regulated protein folding and quality control in the ER. Semin. Cell Dev. Biol. 41, 79–89 (2015).
Sacks, D. et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int. J. Stroke 13, 612–632 (2018).
Watanabe, Y., Bowden, T. A., Wilson, I. A. & Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 1863, 1480–1497 (2019).
Vankadari, N. & Wilce, J. A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microbes Infect. 9, 601–604 (2020).
Lo Presti, A., Rezza, G. & Stefanelli, P. Selective pressure on SARS-CoV-2 protein coding genes and glycosylation site prediction. Heliyon 6, e05001 (2020).
Bagdonaite, I. & Wandall, H. H. Global aspects of viral glycosylation. Glycobiology 28, 443–467 (2018).
Rudd, P. M. et al. Glycosylation and the immune system. Science 291, 2370–2376 (2001).
Chatterjee, S. K., Saha, S. & Munoz, M. N. M. Molecular pathogenesis, immunopathogenesis and novel therapeutic strategy against COVID-19. Front. Mol. Biosci. 7, 196 (2020).
Cipollo, J. F. & Parsons, L. M. Glycomics and glycoproteomics of viruses: Mass spectrometry applications and insights toward structure-function relationships. Mass Spectrom. Rev. 39, 371–409 (2020).
Duan, L. et al. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front. Immunol. 11, 576622 (2020).
Stanley, P. et al. The Consortium of Glycobiology. 99–111 (Cold Spring Harbor Laboratory Press Copyright 2015–2017, 2015).
Nascimento da Silva, L. C. et al. Exploring lectin-glycan interactions to combat COVID-19: lessons acquired from other enveloped viruses. Glycobiology 31, 358–371 (2021).
Hargett, A. A. & Renfrow, M. B. Glycosylation of viral surface proteins probed by mass spectrometry. Curr. Opin. Virol. 36, 56–66 (2019).
Fung, T. S. & Liu, D. X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 13, 405–430 (2018).
Zhang, Y. et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol. Cell. Proteomics 20, 100058 (2020).
Watanabe, Y. et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369, 330–333 (2020).
Wadzinski, T. J. et al. Rapid phenolic O-glycosylation of small molecules and complex unprotected peptides in aqueous solvent. Nat. Chem. 10, 644–652 (2018).
Mayr, J. et al. Unravelling the role of O-glycans in influenza A virus infection. Sci. Rep. 8, 16382 (2018).
Chen, N., Kong, X., Zhao, S. & Xiaofeng, W. Post-translational modification of baculovirus-encoded proteins. Virus Res. 279, 197865 (2020).
Brockhausen, I. et al. The Consortium of Glycobiology (Cold Spring Harbor Laboratory Press Copyright, 2009).
Brockhausen, I. & Stanley, P. in Essentials of Glycobiology (eds. A. Varki et al.) 113–123 (Cold Spring Harbor Laboratory Press, 2015).
Bagdonaite, I. et al. Global mapping of O-glycosylation of varicella zoster virus, human cytomegalovirus, and Epstein-Barr virus. J. Biol. Chem. 291, 12014–12028 (2016).
Shajahan, A., Supekar, N. T., Gleinich, A. S. & Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 30, 981–988 (2020).
Bagdonaite, I. et al. A strategy for O-glycoproteomics of enveloped viruses-the O-glycoproteome of herpes simplex virus type 1. PLoS Pathog. 11, e1004784 (2015).
Xiao, K. J. & Tian, Z. X. GPSeeker enables quantitative structural N-glycoproteomics for site- and structure-specific characterization of differentially expressed N-glycosylation in hepatocellular carcinoma. J. Proteome Res. 18, 2885–2895 (2019).
Wang, Y. & Tian, Z. X. New energy setup strategy for intact N-glycopeptides characterization using higher-energy collisional dissociation. J. Am. Soc. Mass Spectrom. 31, 651–657 (2020).
Wang, Y., Xiao, K. & Tian, Z. Quantitative N-glycoproteomics using stable isotopic diethyl labeling. Talanta 219, 121359 (2020).
Rosenbalm, K. E. et al. Glycomics-informed glycoproteomic analysis of site-specific glycosylation for SARS-CoV-2 spike protein. STAR Protoc. 1, 100214 (2020).
Zhou, D., Xiao, K. & Tian, Z. Separation and detection of minimal length glycopeptide neoantigen epitopes centering the GSTA region of MUC1 by liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 34, e8622 (2020).
Xue, B., Xiao, K., Wang, Y. & Tian, Z. Site- and structure-specific quantitative N-glycoproteomics study of differential N-glycosylation in MCF-7 cancer cells. J. Proteom. 212, 103594 (2020).
Shen, Y., Xiao, K. & Tian, Z. Site- and structure-specific characterization of the human urinary N-glycoproteome with site-determining and structure-diagnostic product ions. Rapid Commun. Mass Spectrom. 35, e8952 (2021).
Xu, F. et al. Quantitative site- and structure-specific N-glycoproteomics characterization of differential N-glycosylation in MCF-7/ADR cancer stem cells. Clin. Proteom. 17, 3 (2020).
Tian, W. et al. O-glycosylation pattern of the SARS-CoV-2 spike protein reveals an “O-Follow-N” rule. Cell Res. 31, 1123–1125 (2021).
Dang, L. et al. Mapping human N-linked glycoproteins and glycosylation sites using mass spectrometry. Trends Anal. Chem. 114, 143–150 (2019).
Hu, H. et al. A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconj. J. 33, 285–296 (2016).
Yu, A. et al. Advances in mass spectrometry-based glycoproteomics. Electrophoresis 39, 3104–3122 (2018).
Gaunitz, S., Nagy, G., Pohl, N. L. & Novotny, M. V. Recent advances in the analysis of complex glycoproteins. Anal. Chem. 89, 389–413 (2017).
Li, X. et al. Databases and bioinformatic tools for glycobiology and glycoproteomics. Int. J. Mol. Sci. 21, 6727 (2020).
Abrahams, J. L. et al. Recent advances in glycoinformatic platforms for glycomics and glycoproteomics. Curr. Opin. Struct. Biol. 62, 56–69 (2020).
Ruhaak, L. R. et al. Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev. 118, 7886–7930 (2018).
Lisacek, F. et al. Databases and associated tools for glycomics and glycoproteomics. Methods Mol. Biol. 1503, 235–264 (2017).
Peng, W. et al. MS-based glycomics and glycoproteomics methods enabling isomeric characterization. Mass Spectrom. Rev. https://doi.org/10.1002/mas.21713 (2021).
Thaysen-Andersen, M. & Packer, N. H. Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 1844, 1437–1452 (2014).
Riley, N. M., Bertozzi, C. R. & Pitteri, S. J. A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Mol. Cell. Proteom. 20, 100029 (2020).
Pujić, I. & Perreault, H. Recent advancements in glycoproteomic studies: Glycopeptide enrichment and derivatization, characterization of glycosylation in SARS CoV2, and interacting glycoproteins. Mass Spectrom. Rev. https://doi.org/10.1002/mas.21679 (2021).
Domann, P. J. et al. Separation-based glycoprofiling approaches using fluorescent labels. Proteomics 7, 70–76 (2007).
Tharmalingam, T. et al. Strategies for the profiling, characterisation and detailed structural analysis of N-linked oligosaccharides. Glycoconj. J. 30, 137–146 (2013).
Pabst, M. & Altmann, F. Glycan analysis by modern instrumental methods. Proteomics 11, 631–643 (2011).
Kolli, V., Schumacher, K. N. & Dodds, E. D. Engaging challenges in glycoproteomics: recent advances in MS-based glycopeptide analysis. Bioanalysis 7, 113–131 (2015).
Wuhrer, M., Catalina, M. I., Deelder, A. M. & Hokke, C. H. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 849, 115–128 (2007).
Desaire, H. Glycopeptide analysis, recent developments and applications. Mol. Cell. Proteom. 12, 893–901 (2013).
Sun, S. & Zhang, H. Identification and validation of atypical N-glycosylation sites. Anal. Chem. 87, 11948–11951 (2015).
Sun, S. et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat. Biotechnol. 34, 84–88 (2016).
Jacobs, J. F., Wevers, R. A., Lefeber, D. J. & van Scherpenzeel, M. Fast, robust and high-resolution glycosylation profiling of intact monoclonal IgG antibodies using nanoLC-chip-QTOF. Clin. Chim. Acta 461, 90–97 (2016).
Haselberg, R., de Jong, G. J. & Somsen, G. W. CE-MS for the analysis of intact proteins 2010-2012. Electrophoresis 34, 99–112 (2013).
Unione, L. et al. Glycoprofile analysis of an intact glycoprotein as inferred by NMR spectroscopy. ACS Cent. Sci. 5, 1554–1561 (2019).
Balaguer, E. & Neusüss, C. Glycoprotein characterization combining intact protein and glycan analysis by capillary electrophoresis-electrospray ionization-mass spectrometry. Anal. Chem. 78, 5384–5393 (2006).
Baerenfaenger, M. & Meyer, B. Intact human alpha-acid glycoprotein analyzed by ESI-qTOF-MS: simultaneous determination of the glycan composition of multiple glycosylation sites. J. Proteome Res. 17, 3693–3703 (2018).
Grunow, D. & Blanchard, V. Enzymatic release of glycoprotein N-glycans and fluorescent labeling. Methods Mol. Biol. 1934, 43–49 (2019).
Tayi, V. S. & Butler, M. Isolation and quantification of N-glycans from immunoglobulin G antibodies for quantitative glycosylation analysis. J. Biol. Methods 2, e19 (2015).
Mamedov, T. et al. A plant-produced in vivo deglycosylated full-length Pfs48/45 as a transmission-blocking vaccine candidate against malaria. Sci. Rep. 9, 1–12 (2019).
Sakayama, K. et al. Glycosylation of lipoprotein lipase in human subcutaneous and omental adipose tissues. Biochim. Biophys. Acta 1127, 153–156 (1992).
Morio, A. et al. Expression, purification, and characterization of highly active endo-α-N-acetylgalactosaminidases expressed by silkworm-baculovirus expression system. J. Asia Pac. Entomol. 22, 404–408 (2019).
Koutsioulis, D., Landry, D. & Guthrie, E. P. J. G. Novel endo-α-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology 18, 799–805 (2008).
Pralow, A. et al. Improvement of the glycoproteomic toolbox with the discovery of a unique C-terminal cleavage specificity of flavastacin for N-glycosylated asparagine. Sci. Rep. 7, 11419 (2017).
Chaze, T. et al. O-glycosylation of the N-terminal region of the serine-rich adhesin Srr1 of Streptococcus agalactiae explored by mass spectrometry. Mol. Cell. Proteomics 13, 2168–2182 (2014).
Bejugam, M., Maltman, B. A. & Flitsch, S. L. J. T. A. Synthesis of N-linked glycopeptides on solid support and their evaluation as protease substrates. Tetrahedron Asymmetry 16, 21–24 (2005).
Vakhrushev, S. Y. et al. Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 12, 932–944 (2013).
Van Rooijen, J. J., Voskamp, A. F., Kamerling, J. P. & Vliegenthart, J. F. J. G. Glycosylation sites and site-specific glycosylation in human Tamm-Horsfall glycoprotein. Glycobiology 9, 21–30 (1999).
Bongers, J. et al. Characterization of glycosylation sites for a recombinant IgG1 monoclonal antibody and a CTLA4-Ig fusion protein by liquid chromatography–mass spectrometry peptide mapping. J. Chromatogr. A 1218, 8140–8149 (2011).
Tumurbaatar, O. & Yoshida, T. Enzymatic digestion and mass spectroscopies of N-linked glycans in lacquer stellacyanin from Rhus vernicifera. Int. J. Polym. Sci. 2015, 547907–547909 (2015).
van der Post, S., Thomsson, K. A. & Hansson, G. C. Multiple enzyme approach for the characterization of glycan modifications on the C-terminus of the intestinal MUC2mucin. J. Proteome Res. 13, 6013–6023 (2014).
Keating, C. L. et al. Spontaneous glycan reattachment following N-glycanase treatment of influenza and HIV vaccine antigens. J. Proteome Res. 19, 733–743 (2020).
Zielinska, D. F., Gnad, F., Wiśniewski, J. R. & Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 (2010).
Lopez Aguilar, A. et al. Tools for studying glycans: recent advances in chemoenzymatic glycan labeling. ACS Chem. Biol. 12, 611–621 (2017).
Yang, S. et al. Deciphering protein O-glycosylation: solid-phase chemoenzymatic cleavage and enrichment. Anal. Chem. 90, 8261–8269 (2018).
Stavenhagen, K., Kolarich, D. & Wuhrer, M. Clinical glycomics employing graphitized carbon liquid chromatography-mass spectrometry. Chromatographia 78, 307–320 (2015).
She, Y. M. et al. Resolving isomeric structures of native glycans by nanoflow porous graphitized carbon chromatography-mass spectrometry. Anal. Chem. 92, 14038–14046 (2020).
Qing, G. et al. Recent advances in hydrophilic interaction liquid interaction chromatography materials for glycopeptide enrichment and glycan separation. TrAC Trends Anal. Chem. 124, 115570 (2019).
Chen, L. et al. Hydrophilic interaction/cation-exchange chromatography for glycopeptide enrichment by using a modified strong-cation exchange material. Anal. Methods 5, 6919–6924 (2013).
Lewandrowski, U. et al. Glycosylation site analysis of human platelets by electrostatic repulsion hydrophilic interaction chromatography. Clin. Proteom. 4, 25–36 (2008).
Zhu, R. et al. Glycoprotein enrichment analytical techniques: advantages and disadvantages. Methods Enzymol. 585, 397–429 (2017).
Zhu, R. et al. Glycoproteins enrichment and LC-MS/MS glycoproteomics in central nervous system applications. Methods Mol. Biol. 1598, 213–227 (2017).
Palmisano, G., Larsen, M. R., Packer, N. H. & Thaysen-Andersen, M. Structural analysis of glycoprotein sialylation – part II: LC-MS based detection. RSC Adv. 3, 22706–22726 (2013).
Hägglund, P. et al. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3, 556–566 (2004).
Merkle, R. K. & Cummings, R. D. Lectin affinity chromatography of glycopeptides. Methods Enzymol. 138, 232–259 (1987).
Lee, W. C. & Lee, K. H. Applications of affinity chromatography in proteomics. Anal. Biochem. 324, 1–10 (2004).
Chen, C. C. et al. Interaction modes and approaches to glycopeptide and glycoprotein enrichment. Analyst 139, 688–704 (2014).
Deeb, S. J., Cox, J., Schmidt-Supprian, M. & Mann, M. N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes. Mol. Cell. Proteom. 13, 240–251 (2014).
Wiśniewski, J. R., Zielinska, D. F. & Mann, M. Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal. Biochem. 410, 307–309 (2011).
Qiu, R. & Regnier, F. E. Use of multidimensional lectin affinity chromatography in differential glycoproteomics. Anal. Chem. 77, 2802–2809 (2005).
Jandera, P. Stationary and mobile phases in hydrophilic interaction chromatography: a review. Anal. Chim. Acta 692, 1–25 (2011).
Lastovickova, M., Strouhalova, D. & Bobalova, J. Use of lectin-based affinity techniques in breast cancer glycoproteomics: a review. J. Proteome Res. 19, 1885–1899 (2020).
Zeng, Z. et al. A proteomics platform combining depletion, multi-lectin affinity chromatography (M-LAC), and isoelectric focusing to study the breast cancer proteome. Anal. Chem. 83, 4845–4854 (2011).
Xu, Y., Zhang, L. & Lu, H. Use of boronic acid nanoparticles in glycoprotein enrichment. Methods Mol. Biol. 951, 45–55 (2013).
Mysling, S., Palmisano, G., Højrup, P. & Thaysen-Andersen, M. Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82, 5598–5609 (2010).
Sun, N. et al. Advances in hydrophilic nanomaterials for glycoproteomics. Chem. Commun. 55, 10359–10375 (2019).
Huang, J. et al. Simultaneous enrichment and separation of neutral and sialyl glycopeptides of SARS-CoV-2 spike protein enabled by dual-functionalized Ti-IMAC material. Anal. Bioanal. Chem. 1–9, https://doi.org/10.1007/s00216-021-03433-1 (2021).
Nilsson, J. et al. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 6, 809–811 (2009).
Huang, T., Armbruster, M. R., Coulton, J. B. & Edwards, J. L. Chemical tagging in mass spectrometry for systems biology. Anal. Chem. 91, 109–125 (2019).
Taga, Y., Kusubata, M., Ogawa-Goto, K. & Hattori, S. Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol. Cell. Proteom. 11, M111.010397 (2012).
Zhang, H., Li, X. J., Martin, D. B. & Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 (2003).
Shi, Z. et al. Boronic acid-modified magnetic Fe(3)O(4)@mTiO(2) microspheres for highly sensitive and selective enrichment of N-glycopeptides in amniotic fluid. Sci. Rep. 7, 4603 (2017).
Bull, S. D. et al. Exploiting the reversible covalent bonding of boronic acids: recognition, sensing, and assembly. Acc. Chem. Res. 46, 312–326 (2013).
Wang, X., Xia, N. & Liu, L. Boronic Acid-based approach for separation and immobilization of glycoproteins and its application in sensing. Int. J. Mol. Sci. 14, 20890–20912 (2013).
Selman, M. H., Hemayatkar, M., Deelder, A. M. & Wuhrer, M. Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal. Chem. 83, 2492–2499 (2011).
Qing, G. et al. Recent advances in hydrophilic interaction liquid interaction chromatography materials for glycopeptide enrichment and glycan separation. TrAC Trends Analyt. Chem. 124, 115570 (2020).
Zhang, C. et al. Evaluation of different N-glycopeptide enrichment methods for N-glycosylation sites mapping in mouse brain. J. Proteome Res. 15, 2960–2968 (2016).
Xiao, H., Chen, W., Smeekens, J. M. & Wu, R. An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins. Nat. Commun. 9, 1692 (2018).
Ongay, S., Boichenko, A., Govorukhina, N. & Bischoff, R. J. J. Glycopeptide enrichment and separation for protein glycosylation analysis. J. Sep. Sci. 35, 2341–2372 (2012).
Neue, K., Mormann, M., Peter-Katalinić, J. & Pohlentz, G. Elucidation of glycoprotein structures by unspecific proteolysis and direct nanoESI mass spectrometric analysis of ZIC-HILIC-enriched glycopeptides. J. Proteome Res. 10, 2248–2260 (2011).
Takegawa, Y. et al. Simple separation of isomeric sialylated N-glycopeptides by a zwitterionic type of hydrophilic interaction chromatography. J. Sep. Sci. 29, 2533–2540 (2006).
Morelle, W. & Michalski, J. C. Analysis of protein glycosylation by mass spectrometry. Nat. Protoc. 2, 1585–1602 (2007).
Stavenhagen, K., Plomp, R. & Wuhrer, M. Site-specific protein N- and O-glycosylation analysis by a C18-porous graphitized carbon-liquid chromatography-electrospray ionization mass spectrometry approach using pronase treated glycopeptides. Anal. Chem. 87, 11691–11699 (2015).
Issaq, H. J. et al. Separation, detection and quantitation of peptides by liquid chromatography and capillary electrochromatography. J. Chromatogr. A 1216, 1825–1837 (2009).
Young, C. et al. In-house packed porous graphitic carbon columns for liquid chromatography-mass spectrometry analysis of N-glycans. Front. Chem. 9, 653959 (2021).
Mechref, Y. et al. Quantitative glycomics strategies. Mol. Cell. Proteom. 12, 874–884 (2013).
Veillon, L. et al. Characterization of isomeric glycan structures by LC-MS/MS. Electrophoresis 38, 2100–2114 (2017).
Vreeker, G. C. & Wuhrer, M. Reversed-phase separation methods for glycan analysis. Anal. Bioanal. Chem. 409, 359–378 (2017).
Gutierrez Reyes, C. D. et al. Glycomics and glycoproteomics: approaches to address isomeric separation of glycans and glycopeptides. J. Sep Sci. 44, 403–425 (2021).
Ji, E. S. et al. Isomer separation of sialylated O- and N-linked glycopeptides using reversed-phase LC-MS/MS at high temperature. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1110–1111, 101–107 (2019).
Hage, D. S. et al. in Liquid Chromatography (Second Edition) (eds Salvatore Fanali, Paul R. Haddad, Colin F. Poole, & Marja-Liisa Riekkola) 319–341 (Elsevier, 2017).
Chirita, R. I. et al. Investigations on the chromatographic behaviour of zwitterionic stationary phases used in hydrophilic interaction chromatography. J. Chromatogr. A 1218, 5939–5963 (2011).
Zauner, G., Koeleman, C. A., Deelder, A. M. & Wuhrer, M. Protein glycosylation analysis by HILIC-LC-MS of Proteinase K-generated N- and O-glycopeptides. J. Sep. Sci. 33, 903–910 (2010).
Pedrali, A. et al. Characterization of intact neo-glycoproteins by hydrophilic interaction liquid chromatography. Molecules 19, 9070–9088 (2014).
Kozlik, P., Goldman, R. & Sanda, M. Hydrophilic interaction liquid chromatography in the separation of glycopeptides and their isomers. Anal. Bioanal. Chem. 410, 5001–5008 (2018).
Royle, L. et al. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem 376, 1–12 (2008).
Guile, G. R. et al. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal. Biochem. 240, 210–226 (1996).
Molnarova, K. & Kozlík, P. J. M. Comparison of different HILIC stationary phases in the separation of hemopexin and immunoglobulin G glycopeptides and their isomers. Molecules 25, 4655 (2020).
Balog, C. I. et al. N-glycosylation of colorectal cancer tissues: a liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics 11, 571–585 (2012).
Reiding, K. R. et al. High-throughput serum N-glycomics: method comparison and application to study rheumatoid arthritis and pregnancy-associated changes*[S]. Mol. Cell. Proteomics 18, 3–15 (2019).
Zhu, R. et al. Isomeric separation of N-glycopeptides derived from glycoproteins by porous graphitic carbon (PGC) LC-MS/MS. Anal. Chem. 92, 9556–9565 (2020).
Stavenhagen, K. et al. N- and O-glycosylation analysis of human C1-inhibitor reveals extensive mucin-type O-glycosylation. Mol. Cell. Proteomics 17, 1225–1238 (2018).
Lohrig, K., Sickmann, A. & Lewandrowski, U. Strong cation exchange chromatography for analysis of sialylated glycopeptides. Methods Mol. Biol. 753, 299–308 (2011).
Zhao, Y. et al. Online coupling of hydrophilic interaction/strong cation exchange/reversed-phase liquid chromatography with porous graphitic carbon liquid chromatography for simultaneous proteomics and N-glycomics analysis. J. Chromatogr. A 1415, 57–66 (2015).
Cai, W. et al. Top-down proteomics of large proteins up to 223 kDa enabled by serial size exclusion chromatography strategy. Anal. Chem. 89, 5467–5475 (2017).
Saraswat, M., Garapati, K., Mun, D. G. & Pandey, A. Extensive heterogeneity of glycopeptides in plasma revealed by deep glycoproteomic analysis using size-exclusion chromatography. Mol. Omics. https://doi.org/10.1039/d1mo00132a (2021).
Alvarez-Manilla, G. et al. Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites. J. Proteome Res. 5, 701–708 (2006).
Kammeijer, G. S. et al. Dopant enriched nitrogen gas combined with sheathless capillary electrophoresis-electrospray ionization-mass spectrometry for improved sensitivity and repeatability in glycopeptide analysis. Anal. Chem. 88, 5849–5856 (2016).
Amon, S., Zamfir, A. D. & Rizzi, A. Glycosylation analysis of glycoproteins and proteoglycans using capillary electrophoresis-mass spectrometry strategies. Electrophoresis 29, 2485–2507 (2008).
Khatri, K. et al. Microfluidic capillary electrophoresis-mass spectrometry for analysis of monosaccharides, oligosaccharides, and glycopeptides. Anal. Chem. 89, 6645–6655 (2017).
An, H. J. & Lebrilla, C. B. Structure elucidation of native N- and O-linked glycans by tandem mass spectrometry (tutorial). Mass Spectrom. Rev. 30, 560–578 (2011).
Scott, N. E. et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, S1–S18 (2011).
Riley, N. M., Malaker, S. A., Driessen, M. D. & Bertozzi, C. R. Optimal dissociation methods differ for N- and O-glycopeptides. J. Proteome Res. 19, 3286–3301 (2020).
Yin, X. et al. Glycoproteomic analysis of the secretome of human endothelial cells. Mol. Cell. Proteomics 12, 956–978 (2013).
Prien, J. M. et al. Differentiating N-linked glycan structural isomers in metastatic and nonmetastatic tumor cells using sequential mass spectrometry. Glycobiology 18, 353–366 (2008).
Shao, W. & Lam, H. Tandem mass spectral libraries of peptides and their roles in proteomics research. Mass Spectrom. Rev. 36, 634–648 (2017).
Mechref, Y., Kang, P. & Novotny, M. V. Differentiating structural isomers of sialylated glycans by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20, 1381–1389 (2006).
Levery, S. B. et al. Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 1850, 33–42 (2015).
Khoo, K. H. Advances toward mapping the full extent of protein site-specific O-GalNAc glycosylation that better reflects underlying glycomic complexity. Curr. Opin. Struct. Biol. 56, 146–154 (2019).
Darula, Z. & Medzihradszky, K. F. Analysis of mammalian O-glycopeptides-we have made a good start, but there is a long way to go. Mol. Cell. Proteom. 17, 2–17 (2018).
Seo, Y., Park, G. M., Oh, M. J. & An, H. J. Investigation of O-glycosylation heterogeneity of recombinant coagulation factor IX using LC-MS/MS. Bioanalysis 9, 1361–1372 (2017).
Hashii, N. & Suzuki, J. Site-specific O-glycosylation analysis by liquid chromatography-mass spectrometry with electron-transfer/higher-energy collisional dissociation. Methods Mol. Biol. 2271, 169–178 (2021).
Wells, L. et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteom. 1, 791–804 (2002).
Liu, D. et al. Site-specific N- and O-glycosylation analysis of human plasma fibronectin. Front. Chem. 9, 691217 (2021).
Hashii, N. & Ishii-Watabe, A. [Site-specific O-glycosylation analysis of therapeutic Fc-fusion protein by mass spectrometry]. Yakugaku Zasshi 138, 1483–1494 (2018).
Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 (2013).
Pasing, Y., Sickmann, A. & Lewandrowski, U. N-glycoproteomics: mass spectrometry-based glycosylation site annotation. Biol. Chem. 393, 249–258 (2012).
Furmanek, A. & Hofsteenge, J. Protein C-mannosylation: facts and questions. Acta Biochim. Pol. 47, 781–789 (2000).
Maynard, J. C., Burlingame, A. L. & Medzihradszky, K. F. Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), a new post-translational modification in mammals. Mol. Cell. Proteom. 15, 3405–3411 (2016).
Hofsteenge, J. et al. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry 33, 13524–13530 (1994).
Schoof, E. M. et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat. Commun. 12, 3341 (2021).
Kolarich, D., Jensen, P. H., Altmann, F. & Packer, N. H. Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 7, 1285–1298 (2012).
Qin, H. et al. Proteomics analysis of site-specific glycoforms by a virtual multistage mass spectrometry method. Anal. Chim. Acta 1070, 60–68 (2019).
Liu, M. et al. Efficient and accurate glycopeptide identification pipeline for high-throughput site-specific N-glycosylation analysis. J. Proteome Res. 13, 3121–3129 (2014).
Kim, U. et al. MS-based technologies for the study of site-specific glycosylation. Mass Spectrom. Lett. 8, 69–78 (2017).
Xiao, K., Shen, Y., Li, S. & Tian, Z. Accurate phosphorylation site localization using phospho-brackets. Anal. Chim. Acta 996, 38–47 (2017).
Harrison, A. G. To b or not to b: the ongoing saga of peptide b ions. Mass Spectrom. Rev. 28, 640–654 (2009).
Reid, G. E., Stephenson, J. L. Jr. & McLuckey, S. A. Tandem mass spectrometry of ribonuclease A and B: N-linked glycosylation site analysis of whole protein ions. Anal. Chem. 74, 577–583 (2002).
Riley, N. M., Hebert, A. S., Westphall, M. S. & Coon, J. J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 10, 1311 (2019).
Zhang, W. et al. Large-scale assignment of N-glycosylation sites using complementary enzymatic deglycosylation. Talanta 85, 499–505 (2011).
Olsen, J. V. et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 (2007).
Wolff, J. J. et al. Electron capture dissociation, electron detachment dissociation and infrared multiphoton dissociation of sucrose octasulfate. Eur. J. Mass Spectrom. 15, 275–281 (2009).
Syka, J. E. et al. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl Acad. Sci. USA 101, 9528–9533 (2004).
Warnke, S. et al. Photodissociation of conformer-selected ubiquitin ions reveals site-specific cis/trans isomerization of proline peptide bonds. J. Am. Chem. Soc. 136, 10308–10314 (2014).
Nilsson, J. Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides. Glycoconj. J. 33, 261–272 (2016).
Yang, Y., Franc, V. & Heck, A. J. R. Glycoproteomics: a balance between high-throughput and in-depth analysis. Trends Biotechnol. 35, 598–609 (2017).
Sanda, M., Benicky, J. & Goldman, R. Low collision energy fragmentation in structure-specific glycoproteomics analysis. Anal. Chem. 92, 8262–8267 (2020).
Zhao, P. et al. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J. Proteome Res. 10, 4088–4104 (2011).
Reiding, K. R., Bondt, A., Franc, V. & Heck, A. J. R. The benefits of hybrid fragmentation methods for glycoproteomics. TrAC Trends Anal. Chem. 108, 260–268 (2018).
Swaney, D. L. et al. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79, 477–485 (2007).
Frese, C. K. et al. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84, 9668–9673 (2012).
Kronewitter, S. R. et al. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics 9, 2986–2994 (2009).
Solá, R. J. & Griebenow, K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 98, 1223–1245 (2009).
Haltiwanger, R. S. & Lowe, J. B. Role of glycosylation in development. Annu. Rev. Biochem 73, 491–537 (2004).
Moremen, K. W., Tiemeyer, M. & Nairn, A. V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 (2012).
Chandler, K. B. & Costello, C. E. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis 37, 1407–1419 (2016).
Čaval, T., Heck, A. J. R. & Reiding, K. R. Meta-heterogeneity: evaluating and describing the diversity in glycosylation between sites on the same glycoprotein. Mol. Cell. Proteom. 20, 100010 (2020).
Khatri, K. et al. Integrated omics and computational glycobiology reveal structural basis for influenza a virus glycan microheterogeneity and host interactions. Mol. Cell. Proteom. 15, 1895–1912 (2016).
Dennis, J. W. et al. Beta 1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science 236, 582–585 (1987).
Gabius, H. J. et al. From lectin structure to functional glycomics: principles of the sugar code. Trends Biochem. Sci. 36, 298–313 (2011).
Freeze, H. H. Understanding human glycosylation disorders: biochemistry leads the charge. J. Biol. Chem. 288, 6936–6945 (2013).
Huang, Y. et al. FUT8-mediated aberrant N-glycosylation of B7H3 suppresses the immune response in triple-negative breast cancer. Nat. Commun. 12, 2672 (2021).
Croci, D. O. et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell 156, 744–758 (2014).
Ardejani, M. S., Noodleman, L., Powers, E. T. & Kelly, J. W. Stereoelectronic effects in stabilizing protein–N-glycan interactions revealed by experiment and machine learning. Nat. Chem. 13, 480–487 (2021).
Park, J. J. & Lee, M. Increasing the α 2, 6 sialylation of glycoproteins may contribute to metastatic spread and therapeutic resistance in colorectal cancer. Gut Liver 7, 629–641 (2013).
Strasser, R. Biological significance of complex N-glycans in plants and their impact on plant physiology. Front. Plant Sci. 5, 363 (2014).
Clerc, F. et al. Human plasma protein N-glycosylation. Glycoconj. J. 33, 309–343 (2016).
Liew, C. Y. et al. Structural identification of N-glycan isomers using logically derived sequence tandem mass spectrometry. Commun. Chem. 4, 92 (2021).
Chernykh, A., Kawahara, R. & Thaysen-Andersen, M. Towards structure-focused glycoproteomics. Biochem. Soc. Trans. 49, 161–186 (2021).
Shen, J. et al. StrucGP: de novo structural sequencing of site-specific N-glycan on glycoproteins using a modularization strategy. Nat. Methods 18, 921–929 (2021).
Gindzienska-Sieskiewicz, E. et al. The changes in monosaccharide composition of immunoglobulin G in the course of rheumatoid arthritis. Clin. Rheumatol. 26, 685–690 (2007).
Stanley, P. A method to the madness of N-glycan complexity? Cell 129, 27–29 (2007).
Cai, X. et al. The importance of N-glycosylation on β(3) integrin ligand binding and conformational regulation. Sci. Rep. 7, 4656 (2017).
Xiao, K. et al. Large-scale identification and visualization of N-glycans with primary structures using GlySeeker. Rapid Commun. Mass Spectrom. 32, 142–148 (2018).
Maass, K. et al. “Glyco-peakfinder”-de novo composition analysis of glycoconjugates. Proteomics 7, 4435–4444 (2007).
Haslam, S. M. et al. Glycoinformatics for Structural Glycomics. Glyco-Bioinformatics-Bits “n” Bytes of Sugars (ISBN: 978-3-8325-2719-8). Chapter 1 (2011).
Xiao, K. & Tian, Z. Site- and structure-specific quantitative N-glycoproteomics using RPLC-pentaHILIC separation and the intact N-glycopeptide search engine GPSeeker. Curr. Protoc. Protein Sci. 97, e94 (2019).
Pan, K. T., Chen, C. C., Urlaub, H. & Khoo, K. H. Adapting data-independent acquisition for mass spectrometry-based protein site-specific N-glycosylation analysis. Anal. Chem. 89, 4532–4539 (2017).
Cao, L. et al. Global site-specific analysis of glycoprotein N-glycan processing. Nat. Protoc. 13, 1196–1212 (2018).
Yang, Y. et al. GproDIA enables data-independent acquisition glycoproteomics with comprehensive statistical control. Nat. Commun. 12, 6073 (2021).
Pompach, P. et al. Semi-automated identification of N-Glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase LC-MS/MS, and glycan database search. J. Proteome Res. 11, 1728–1740 (2012).
Choo, M. S., Wan, C., Rudd, P. M. & Nguyen-Khuong, T. GlycopeptideGraphMS: improved glycopeptide detection and identification by exploiting graph theoretical patterns in mass and retention time. Anal. Chem. 91, 7236–7244 (2019).
Woodin, C. L. et al. GlycoPep grader: a web-based utility for assigning the composition of N-linked glycopeptides. Anal. Chem. 84, 4821–4829 (2012).
Liang, S. Y. et al. An adaptive workflow coupled with Random Forest algorithm to identify intact N-glycopeptides detected from mass spectrometry. Bioinformatics 30, 1908–1916 (2014).
Lynn, K. S. et al. MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS² approach. Anal. Chem. 87, 2466–2473 (2015).
Park, G. W. et al. Integrated GlycoProteome Analyzer (I-GPA) for automated identification and quantitation of site-specific N-glycosylation. Sci. Rep. 6, 21175 (2016).
Cao, L. et al. Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation. Anal. Biochem. 452, 96–102 (2014).
Mao, J. et al. A new searching strategy for the identification of O-linked glycopeptides. Anal. Chem. 91, 3852–3859 (2019).
Schulze, S. et al. SugarPy facilitates the universal, discovery-driven analysis of intact glycopeptides. Bioinformatics btaa1042, https://doi.org/10.1093/bioinformatics/btaa1042 (2020).
Campbell, M. P. et al. UniCarbKB: putting the pieces together for glycomics research. Proteomics 11, 4117–4121 (2011).
Lütteke, T. et al. GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology 16, 71r–81r (2006).
Cooper, C. A., Harrison, M. J., Wilkins, M. R. & Packer, N. H. GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 29, 332–335 (2001).
Cooper, C. A. et al. GlycoSuiteDB: a curated relational database of glycoprotein glycan structures and their biological sources. 2003 update. Nucleic Acids Res. 31, 511–513 (2003).
Campbell, M. P. & Packer, N. H. UniCarbKB: new database features for integrating glycan structure abundance, compositional glycoproteomics data, and disease associations. Biochim. Biophys. Acta 1860, 1669–1675 (2016).
Campbell, M. P. et al. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 42, D215–D221 (2014).
von der Lieth, C. W. et al. EUROCarbDB: an open-access platform for glycoinformatics. Glycobiology 21, 493–502 (2011).
Ahmad Izaham, A. R. & Scott, N. E. Open database searching enables the identification and comparison of bacterial glycoproteomes without defining glycan compositions prior to searching. Mol. Cell. Proteom. 19, 1561–1574 (2020).
Zhao, S. et al. GlycoStore: a database of retention properties for glycan analysis. Bioinformatics 34, 3231–3232 (2018).
Aoki-Kinoshita, K. et al. GlyTouCan 1.0-The international glycan structure repository. Nucleic Acids Res. 44, D1237–D1242 (2016).
Damerell, D. et al. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol. Chem. 393, 1357–1362 (2012).
Zhu, Z. et al. GlycoPep Detector: a tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra. Anal. Chem. 85, 5023–5032 (2013).
Go, E. P. et al. GlycoPep DB: a tool for glycopeptide analysis using a “Smart Search”. Anal. Chem. 79, 1708–1713 (2007).
Chang, D. & Zaia, J. Methods to improve quantitative glycoprotein coverage from bottom-up LC-MS data. Mass Spectrom. Rev. https://doi.org/10.1002/mas.21692 (2021).
Riley, N. M., Malaker, S. A., Driessen, M. D. & Bertozzi, C. R. Optimal dissociation methods differ for N- and O-glycopeptides. J. Proteome Res. 19, 3286–3301 (2020).
McCallum, M. et al. Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation. Nat. Struct. Mol. Biol. 27, 942–949 (2020).
Bern, M., Cai, Y. & Goldberg, D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 79, 1393–1400 (2007).
Zeng, W. F. et al. pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Sci. Rep. 6, 25102 (2016).
Zeng, W.-F. et al. Precise, fast and comprehensive analysis of intact glycopeptides and monosaccharide-modifications with pGlyco3. bioRxiv https://doi.org/10.1101/2021.02.06.430063 (2021).
Toghi Eshghi, S. et al. GPQuest: a spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides. Anal. Chem. 87, 5181–5188 (2015).
Lu, L. et al. O-pair search with MetaMorpheus for O-glycopeptide characterization. Nat. Methods 17, 1133–1138 (2020).
Polasky, D. A., Yu, F., Teo, G. C. & Nesvizhskii, A. I. Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco. Nat. Methods 17, 1125–1132 (2020).
Pai, P. J., Hu, Y. & Lam, H. Direct glycan structure determination of intact N-linked glycopeptides by low-energy collision-induced dissociation tandem mass spectrometry and predicted spectral library searching. Anal. Chim. Acta 934, 152–162 (2016).
Zhu, J. et al. Differential quantitative determination of site-specific intact N-glycopeptides in serum haptoglobin between hepatocellular carcinoma and cirrhosis using LC-EThcD-MS/MS. J. Proteome Res. 18, 359–371 (2019).
Giménez, E., Gay, M. & Vilaseca, M. Automatic and rapid identification of glycopeptides by nano-UPLC-LTQ-FT-MS and proteomic search engine. J. Proteom. 152, 236–242 (2017).
Yu, Z. et al. Sequential fragment ion filtering and endoglycosidase-assisted identification of intact glycopeptides. Anal. Bioanal. Chem. 409, 3077–3087 (2017).
Bern, M., Kil, Y. J. & Becker, C. Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinform. Chapter 13, Unit13.20 (2012).
Ye, Z. & Vakhrushev, S. Y. The role of data-independent acquisition for glycoproteomics. Mol. Cell Proteom. 20, 100042 (2021).
Ye, Z., Mao, Y., Clausen, H. & Vakhrushev, S. Y. Glyco-DIA: a method for quantitative O-glycoproteomics with in silico-boosted glycopeptide libraries. Nat. Methods 16, 902–910 (2019).
Sinitcyn, P. et al. MaxDIA enables library-based and library-free data-independent acquisition proteomics. Nat Biotechnol https://doi.org/10.1038/s41587-021-00968-7 (2021).
Xu, L. L., Young, A., Zhou, A. & Röst, H. L. Machine learning in mass spectrometric analysis of DIA data. Proteomics 20, e1900352 (2020).
Lin, C. H., Krisp, C., Packer, N. H. & Molloy, M. P. Development of a data independent acquisition mass spectrometry workflow to enable glycopeptide analysis without predefined glycan compositional knowledge. J. Proteom. 172, 68–75 (2018).
Phung, T. K., Zacchi, L. F. & Schulz, B. L. DIALib: an automated ion library generator for data independent acquisition mass spectrometry analysis of peptides and glycopeptides. Mol. Omics 16, 100–112 (2020).
Yang, X. et al. Proteome-wide analysis of N-glycosylation stoichiometry using SWATH technology. J. Proteome Res. 16, 3830–3840 (2017).
Yuan, W. et al. Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LC-MS-MRM in liver disease. J. Proteom. 116, 24–33 (2015).
Zacchi, L. F. & Schulz, B. L. SWATH-MS glycoproteomics reveals consequences of defects in the glycosylation machinery. Mol. Cell. Proteom. 15, 2435–2447 (2016).
Roth, Z., Yehezkel, G. & Khalaila, I. Identification and quantification of protein glycosylation. Int. J. Carbohydr. Chem. 2012, 640923 (2012).
Dimitrov, D. S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2, 109–122 (2004).
Li, Y. et al. The importance of glycans of viral and host proteins in enveloped virus infection. Front. Immunol. 12, 638573 (2021).
Vigerust, D. J. Protein glycosylation in infectious disease pathobiology and treatment. Cent. Eur. J. Biol. 6, 802–816 (2011).
Alexandersen, S., Chamings, A. & Bhatta, T. R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 11, 6059 (2020).
Malik, Y. A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 42, 3–11 (2020).
Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e910 (2020).
Gordon, D. E. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020).
Fehr, A. R. & Perlman, S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23 (2015).
Chan, J. F. et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 9, 221–236 (2020).
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Kumar, S., Nyodu, R., Maurya, V. K. & Saxena, S. K. J. C. D. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Coronavirus Disease 2019 (COVID-19) 23 (2020).
Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860 (2020).
Xu, X. et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 63, 457–460 (2020).
Archambault, A. N. et al. Cumulative burden of colorectal cancer-associated genetic variants is more strongly associated with early-onset vs late-onset cancer. Gastroenterology 158, 1274–1286.e1212 (2020).
Zhang, Y. & Kutateladze, T. G. Molecular structure analyses suggest strategies to therapeutically target SARS-CoV-2. Nat. Commun. 11, 2920 (2020).
Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e286 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e278 (2020).
Huang, Y. et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149 (2020).
Shang, J. et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020).
Zhang, H. et al. Expression of the SARS-CoV-2 ACE2 Receptor in the human airway epithelium. Am. J. Respir. Crit. Care Med. 202, 219–229 (2020).
Hasöksüz, M., Kiliç, S. & Saraç, F. Coronaviruses and SARS-COV-2. Turk. J. Med. Sci. 50, 549–556 (2020).
Samrat, S. K., Tharappel, A. M., Li, Z. & Li, H. Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res. 288, 198141 (2020).
Walls, A. C. et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039.e1015 (2019).
Antonopoulos, A. et al. Site-specific characterization of SARS-CoV-2 spike glycoprotein receptor-binding domain. Glycobiology 31, 181–187 (2021).
Wang, D. et al. Comprehensive analysis of the glycan complement of SARS-CoV-2 spike proteins using signature ions-triggered electron-transfer/higher-energy collisional dissociation (EThcD) mass spectrometry. Anal. Chem. 92, 14730–14739 (2020).
Lenza, M. P. et al. Structural characterization of n-linked glycans in the receptor binding domain of the SARS-CoV-2 spike protein and their interactions with human lectins. Angew. Chem. Int. Ed. Engl. 59, 23763–23771 (2020).
Yao, H. et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 183, 730–738.e713 (2020).
Sanda, M., Morrison, L. & Goldman, R. N- and O-Glycosylation of the SARS-CoV-2 Spike Protein. Anal. Chem. 93, 2003–2009 (2021).
Yang, J. et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586, 572–577 (2020).
Zhou, D. et al. Identification of 22 N-glycosites on spike glycoprotein of SARS-CoV-2 and accessible surface glycopeptide motifs: implications for vaccination and antibody therapeutics. Glycobiology 31, 69–80 (2021).
Miller, L. M. et al. Heterogeneity of glycan processing on trimeric SARS-CoV-2 spike protein revealed by charge detection mass spectrometry. J. Am. Chem. Soc. 143, 3959–3966 (2021).
Bagdonaite, I. et al. Site-specific O-glycosylation analysis of SARS-CoV-2 spike protein produced in insect and human cells. Viruses 13, 551 (2021).
Andersen, K. G. et al. The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020).
Dong, X. et al. Comprehensive O-glycosylation analysis of the SARS-CoV-2 spike protein with biomimetic Trp-Arg materials. Anal. Chem. 93, 10444–10452 (2021).
Zhang, Y. et al. Mucin-type O-glycosylation landscapes of SARS-CoV-2 Spike Proteins. bioRxiv https://doi.org/10.1101/2020.07.29.227785 (2020).
Roberts, D. S. et al. Structural O-glycoform heterogeneity of the SARS-CoV-2 spike protein receptor-binding domain revealed by top-down mass spectrometry. J. Am. Chem. Soc. 143, 12014–12024 (2021).
Gstöttner, C. et al. Structural and functional characterization of SARS-CoV-2 RBD domains produced in mammalian cells. Anal. Chem. 93, 6839–6847 (2021).
Wang, Y. et al. Impact of expressing cells on glycosylation and glycan of the SARS-CoV-2 spike glycoprotein. ACS Omega 6, 15988–15999 (2021).
Brun, J. et al. Assessing antigen structural integrity through glycosylation analysis of the SARS-CoV-2 viral spike. ACS Cent. Sci. 7, 586–593 (2021).
Lowenthal, M. S. et al. Identification of novel N-glycosylation sites at noncanonical protein consensus motifs. J. Proteome Res. 15, 2087–2101 (2016).
Shajahan, A. et al. Comprehensive characterization of N- and O- glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology 31, 410–424 (2021).
Zhang, S. et al. Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution. Nat. Commun. 12, 1607 (2021).
Sztain, T. et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. bioRxiv https://doi.org/10.1101/2021.02.15.431212 (2021).
Cao, Y. et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84.e16 (2020).
Winstone, H. et al. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol. 95, e02422–20 (2021).
Jaimes, J. A., Millet, J. K. & Whittaker, G. R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 23, 101212 (2020).
Johnson, B. A. et al. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv https://doi.org/10.1101/2020.08.26.268854 (2020).
Nao, N. et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. mBio. 8, e02298–16 (2017).
Teng, S. et al. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 22, 1239–1253 (2021).
Rahnama, S., Azimzadeh Irani, M., Amininasab, M. & Ejtehadi, M. R. S494 O-glycosylation site on the SARS-CoV-2 RBD affects the virus affinity to ACE2 and its infectivity; a molecular dynamics study. Sci. Rep. 11, 15162 (2021).
Zhang, Y. et al. O-glycosylation landscapes of SARS-CoV-2 spike proteins. Front. Chem. 9, 689521 (2021).
Zhang, L. et al. O-glycosylation of the novel SARS-CoV-2 coronavirus spike protein influences furin cleavage. FASEB J. 35. https://doi.org/10.1096/fasebj.2021.35.S1.00261 (2021).
Zhang, L. et al. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. bioRxiv https://doi.org/10.1101/2021.02.05.429982 (2021).
Schjoldager, K. T. et al. A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J. Biol. Chem. 286, 40122–40132 (2011).
Uslupehlivan, M. & Şener, E. Glycoinformatics approach for identifying target positions to inhibit initial binding of SARS-CoV-2 S1 protein to the host cell. bioRxiv https://doi.org/10.1101/2020.03.25.007898 (2020).
McClenaghan, C., Hanson, A., Lee, S. J. & Nichols, C. G. Coronavirus proteins as ion channels: current and potential research. Front. Immunol. 11, 573339 (2020).
Schoeman, D. & Fielding, B. C. Coronavirus envelope protein: current knowledge. Virol. J. 16, 69 (2019).
Artika, I. M., Dewantari, A. K. & Wiyatno, A. Molecular biology of coronaviruses: current knowledge. Heliyon 6, e04743 (2020).
Park, S. H. et al. Interactions of SARS-CoV-2 envelope protein with amilorides correlate with antiviral activity. PLoS Pathog. 17, e1009519 (2021).
Mandala, V. S. et al. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 27, 1202–1208 (2020).
Sarkar, M. & Saha, S. Structural insight into the role of novel SARS-CoV-2 E protein: a potential target for vaccine development and other therapeutic strategies. PLoS ONE 15, e0237300 (2020).
Pervushin, K. et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 5, e1000511 (2009).
Wilson, L., Gage, P. & Ewart, G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353, 294–306 (2006).
Surya, W. et al. MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res. 201, 61–66 (2015).
Verdiá-Báguena, C. et al. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology 432, 485–494 (2012).
Nieto-Torres, J. L. et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 10, e1004077 (2014).
Ruch, T. R. & Machamer, C. E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 85, 675–685 (2011).
Singh Tomar, P. P. & Arkin, I. T. SARS-CoV-2 E protein is a potential ion channel that can be inhibited by Gliclazide and Memantine. Biochem. Biophys. Res. Commun. 530, 10–14 (2020).
Duart, G. et al. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 10, 200209 (2020).
Thomas, S. The structure of the membrane protein of SARS-CoV-2 resembles the sugar transporter semiSWEET. Pathog. Immun. 5, 342–363 (2020).
Fu, Y. Z. et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol. Immunol. 18, 613–620 (2021).
Boson, B. et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 296, 100111 (2021).
Masters, P. S. The molecular biology of coronaviruses. Adv. Virus Res. 66, 193–292 (2006).
de Haan, C. A. & Rottier, P. J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64, 165–230 (2005).
Dawood, A. A. Glycosylation, ligand binding sites and antigenic variations between membrane glycoprotein of COVID-19 and related coronaviruses. Vacunas 22, 1–9 (2021).
Lu, R. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 (2020).
Liu, D. X. et al. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 109, 97–109 (2014).
Majumdar, P. & Niyogi, S. ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 148, e262 (2020).
Ren, Y. et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 17, 881–883 (2020).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020).
Oostra, M., de Haan, C. A., de Groot, R. J. & Rottier, P. J. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 80, 2326–2336 (2006).
Yadav, R. et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 10, 821 (2021).
Samadizadeh, S. et al. COVID-19: Why does disease severity vary among individuals? Respir. Med. 180, 106356 (2021).
Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).
Chen, M. et al. An epigenetic mechanism underlying chromosome 17p deletion-driven tumorigenesis. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-20-0336 (2020).
South, A. M., Diz, D. I. & Chappell, M. C. COVID-19, ACE2, and the cardiovascular consequences. Am. J. Physiol. Heart Circ. Physiol. 318, H1084–h1090 (2020).
Nishiga, M. et al. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 17, 543–558 (2020).
Allen, J. D. et al. Subtle Influence of ACE2 Glycan Processing on SARS-CoV-2 Recognition. J. Mol. Biol. 433, 166762 (2021).
Li, W. et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl Acad. Sci. USA 114, E8508–e8517 (2017).
Schwegmann-Wessels, C. et al. The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus. Virol. J. 8, 435 (2011).
Tortorici, M. A. et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 26, 481–489 (2019).
Wielgat, P., Rogowski, K., Godlewska, K. & Car, H. Coronaviruses: is sialic acid a gate to the eye of cytokine storm? From the entry to the effects. Cells 9, 1963 (2020).
Huang, X. et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 89, 7202–7213 (2015).
Chu, H. et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 12, 134 (2021).
Procko, E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. bioRxiv https://doi.org/10.1101/2020.03.16.994236 (2020).
Mehdipour, A. R. & Hummer, G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl Acad. Sci. USA 118, e2100425118 (2021).
Khoury, D. S. et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021).
Edara, V. V. et al. Neutralizing antibodies against SARS-CoV-2 variants after infection and vaccination. JAMA 325, 1896–1898 (2021).
Mellet, J. & Pepper, M. S. A COVID-19 vaccine: big strides come with big challenges. Vaccines 9, 39 (2021).
Dispinseri, S. et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 12, 2670 (2021).
Tortorici, M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Ledford, H. COVID antibody treatments show promise for preventing severe disease. Nature 591, 513–514 (2021).
Cathcart, A. L. et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv https://doi.org/10.1101/2021.03.09.434607 (2021).
Holodick, N. E., Rodríguez-Zhurbenko, N. & Hernández, A. M. Defining natural antibodies. Front. Immunol. 8, 872 (2017).
Palma, J., Tokarz-Deptuła, B., Deptuła, J. & Deptuła, W. Natural antibodies - facts known and unknown. Cent. Eur. J. Immunol. 43, 466–475 (2018).
Lardone, R. D. et al. How glycobiology can help us treat and beat the COVID-19 pandemic. J. Biol. Chem. 296, 100375 (2021).
Zlocowski, N. et al. Purified human anti-Tn and anti-T antibodies specifically recognize carcinoma tissues. Sci. Rep. 9, 8097 (2019).
Cornelissen, L. A. M. et al. Tn antigen expression contributes to an immune suppressive microenvironment and drives tumor growth in colorectal cancer. Front. Oncol. 10, 1622 (2020).
Breiman, A. et al. Low levels of natural anti-α-N-acetylgalactosamine (Tn) antibodies are associated with COVID-19. Front. Microbiol. 12, 641460 (2021).
Williams, W. B. et al. Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Cell 184, 2955–2972.e2925 (2021).
Dai, L. & Gao, G. F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 21, 73–82 (2021).
Gao, Q. et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81 (2020).
Han, B. et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy children and adolescents: a double-blind, randomised, controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 21, 803–812 (2021).
Jara, A. et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. New Engl. J. Med. 385, 946–948 (2021).
Yu, J. et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020).
Nishikawa, T. et al. Anti-CoVid19 plasmid DNA vaccine induces a potent immune response in rodents by Pyro-drive Jet Injector intradermal inoculation. bioRxiv https://doi.org/10.1101/2021.01.13.426436 (2021).
van Doremalen, N. et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582 (2020).
Buchbinder, S. P., McElrath, M. J., Dieffenbach, C. & Corey, L. Use of adenovirus type-5 vectored vaccines: a cautionary tale. Lancet 396, e68–e69 (2020).
Corbett, K. S. et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. New Engl. J. Med. 383, 1544–1555 (2020).
Jackson, L. A. et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. New Engl. J. Med. 383, 1920–1931 (2020).
Keech, C. et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. New Engl. J. Med. 383, 2320–2332 (2020).
Dong, Y. et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target Ther. 5, 237 (2020).
Krammer, F. SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020).
Tan, H. X. et al. Immunogenicity of prime-boost protein subunit vaccine strategies against SARS-CoV-2 in mice and macaques. Nat. Commun. 12, 1403 (2021).
Ravichandran, S. et al. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 12, eabc3539 (2020).
Watanabe, Y. et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 11, 2688 (2020).
Doores, K. J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 282, 4679–4691 (2015).
Balzarini, J. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 5, 583–597 (2007).
Wang, D. Coronaviruses’ sugar shields as vaccine candidates. Curr. Trends Immunol. 21, 17–23 (2020).
Gao, C. et al. SARS-CoV-2 spike protein interacts with multiple innate immune receptors. bioRxiv https://doi.org/10.1101/2020.07.29.227462 (2020).
de Oliveira Figueiroa, E. et al. Lectin-carbohydrate interactions: implications for the development of new anticancer agents. Curr. Med. Chem. 24, 3667–3680 (2017).
Liu, Y. et al. The roles of direct recognition by animal lectins in antiviral immunity and viral pathogenesis. Molecules 20, 2272–2295 (2015).
Liu, Y. M. et al. A carbohydrate-binding protein from the edible lablab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell Rep. 32, 108016 (2020).
Mori, T. et al. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 280, 9345–9353 (2005).
Sharma, V. K., Sharma, I. & Glick, J. The expanding role of mass spectrometry in the field of vaccine development. Mass Spectrom. Rev. 39, 83–104 (2020).
Cai, Y. et al. Griffithsin with a broad-spectrum antiviral activity by binding glycans in viral glycoprotein exhibits strong synergistic effect in combination with a pan-coronavirus fusion inhibitor targeting SARS-CoV-2 spike S2 subunit. Virol. Sin. 35, 857–860 (2020).
Hoffmann, D. et al. Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites. EMBO J. 40, e108375 (2021).
Wang, W. et al. Lentil lectin derived from Lens culinaris exhibit broad antiviral activities against SARS-CoV-2 variants. Emerg. Microbes Infect. 10, 1519–1529 (2021).
Fajgenbaum, D. C. & June, C. H. Cytokine storm. New Engl. J. Med. 383, 2255–2273 (2020).
Song, P. et al. Cytokine storm induced by SARS-CoV-2. Clin. Chim. Acta 509, 280–287 (2020).
Tang, Y. et al. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 11, 1708 (2020).
Coperchini, F. et al. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 53, 25–32 (2020).
Talaga, M. L. et al. Multitasking human lectin galectin-3 interacts with sulfated glycosaminoglycans and chondroitin sulfate proteoglycans. Biochemistry 55, 4541–4551 (2016).
Wang, W. H. et al. The role of galectins in virus infection - a systemic literature review. J. Microbiol. Immunol. Infect. 53, 925–935 (2020).
Caniglia, J. L. et al. Immunopathology of galectin-3: an increasingly promising target in COVID-19. F1000Research 9, 1078 (2020).
Garcia-Revilla, J., Deierborg, T., Venero, J. L. & Boza-Serrano, A. Hyperinflammation and fibrosis in severe COVID-19 patients: galectin-3, a target molecule to consider. Front. Immunol. 11, 2069 (2020).
Caniglia, J. L. et al. A potential role for Galectin-3 inhibitors in the treatment of COVID-19. PeerJ 8, e9392 (2020).
Saito, S. et al. Pharmacotherapy and adjunctive treatment for idiopathic pulmonary fibrosis (IPF). J. Thorac. Dis. 11, S1740–s1754 (2019).
Ren, Z. et al. Gal-3 is a potential biomarker for spinal cord injury and Gal-3 deficiency attenuates neuroinflammation through ROS/TXNIP/NLRP3 signaling pathway. Biosci. Rep. 39, BSR20192368 (2019).
Chalasani, N. et al. Effects of belapectin, an inhibitor of galectin-3, in patients with nonalcoholic steatohepatitis with cirrhosis and portal hypertension. Gastroenterology 158, 1334–1345.e1335 (2020).
Williams, S. J. & Goddard-Borger, E. D. α-glucosidase inhibitors as host-directed antiviral agents with potential for the treatment of COVID-19. Biochem. Soc. Trans. 48, 1287–1295 (2020).
Evans DeWald, L. et al. Iminosugars: a host-targeted approach to combat Flaviviridae infections. Antivir. Res. 184, 104881 (2020).
O’Keefe, S. et al. Characterizing the selectivity of ER α-glucosidase inhibitors. Glycobiology 29, 530–542 (2019).
Clarke, E. C., Nofchissey, R. A., Ye, C. & Bradfute, S. B. The iminosugars celgosivir, castanospermine and UV-4 inhibit SARS-CoV-2 replication. Glycobiology 31, 378–384 (2021).
Rajasekharan, S. et al. Inhibitors of protein glycosylation are active against the coronavirus severe acute respiratory syndrome coronavirus SARS-CoV-2. Viruses 13, 808 (2021).
Elbein, A. D., Tropea, J. E., Mitchell, M. & Kaushal, G. P. Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J. Biol. Chem. 265, 15599–15605 (1990).
Nicholls, J. M., Moss, R. B. & Haslam, S. M. The use of sialidase therapy for respiratory viral infections. Antivir. Res. 98, 401–409 (2013).
Triana-Baltzer, G. B. et al. DAS181, a sialidase fusion protein, protects human airway epithelium against influenza virus infection: an in vitro pharmacodynamic analysis. J. Antimicrob. Chemother. 65, 275–284 (2010).
Banerjee, N. & Mukhopadhyay, S. Viral glycoproteins: biological role and application in diagnosis. Virusdisease 27, 1–11 (2016).
Supekar, N. T. et al. Variable post-translational modifications of SARS-CoV-2 nucleocapsid protein. Glycobiology 31, 1080–1092 (2021).
Tipnis, S. R. et al. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275, 33238–33243 (2000).
van der Laarse, S. A. M., Leney, A. C. & Heck, A. J. R. Crosstalk between phosphorylation and O-GlcNAcylation: friend or foe. FEBS J. 285, 3152–3167 (2018).
Pandeswari, P. B. & Sabareesh, V. Middle-down approach: a choice to sequence and characterize proteins/proteomes by mass spectrometry. RSC Adv. 9, 313–344 (2019).
Yang, Y. et al. Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat. Commun. 7, 13397 (2016).
Volz, E. et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184, 64–75.e11 (2021).
Korber, B. et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182, 812–827.e819 (2020).
Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020).
Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e1220 (2020).
Thomson, E. C. et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e1120 (2021).
Lopez Bernal, J. et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. New Engl. J. Med. 385, 585–594 (2021).
Planas, D. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021).
Singh, J. et al. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 27, 1131–1133 (2021).
Acknowledgements
This work was supported by the National Key R&D Program of China (No. 2018YFC2000305 and 2018YFC1312301), National Natural Science Foundation of China (No. 81570060, 31870826, 82073221, 21775110, and 22074105), the program of National Clinical Research Center for Geriatrics of West China Hospital (No. Z20191001), West China Hospital 1.3.5 project for disciplines of excellence (No. ZYYC20007), and Shanghai Science and Technology Commission (14DZ2261100).
Author information
Authors and Affiliations
Contributions
Z.T. and L.D. designed the outline and revised the manuscript. Y.G and S.Q. drafted the manuscript. All authors have read and approved the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gong, Y., Qin, S., Dai, L. et al. The glycosylation in SARS-CoV-2 and its receptor ACE2. Sig Transduct Target Ther 6, 396 (2021). https://doi.org/10.1038/s41392-021-00809-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41392-021-00809-8
This article is cited by
-
COVID-19 immune signatures in Uganda persist in HIV co-infection and diverge by pandemic phase
Nature Communications (2024)
-
High-throughput screening of spike variants uncovers the key residues that alter the affinity and antigenicity of SARS-CoV-2
Cell Discovery (2023)
-
Association between soluble angiotensin-converting enzyme 2 in saliva and SARS-CoV-2 infection: a cross-sectional study
Scientific Reports (2023)
-
Development of an integrated and inferenceable RDF database of glycan, pathogen and disease resources
Scientific Data (2023)
-
Analysis of the N-glycosylation profiles of the spike proteins from the Alpha, Beta, Gamma, and Delta variants of SARS-CoV-2
Analytical and Bioanalytical Chemistry (2023)