Abstract
Background
Previous studies have related FAM3C gene with childhood bone health, and the regulation of lipid metabolism in hepatocytes. The present case−control study aimed to analyze the association of FAM3C genetic variants with overweight/obesity and lipid traits among Chinese children.
Methods
Two genetic variants (rs7776725 and rs7793554) within the FAM3C gene were genotyped in 3305 Chinese children aged 6–18 years.
Results
In the whole study population, the T-allele of rs7776725 and A-allele of rs7793554 within the FAM3C gene were associated with 40.2% (95% CI: 11.6−76.1%; P = 0.004) and 29.1% (6.9−56.0%; P = 0.008) increased risk of dyslipidemia, higher triglyceride (P = 0.014 and P = 0.001) and lower HDL-C (P = 0.015 and P = 0.003). In addition, we found that rs7776725 interacted with sex on dyslipidemia (Pfor interaction = 0.004), and sex-stratified analyses showed that it was significantly associated with dyslipidemia only in girls (P = 8.78 × 10–5). The variant also showed nominally significant interactions with sex on total cholesterol and LDL-C (Pfor interaction = 0.012 and 0.008).
Conclusion
We found that FAM3C genetic variants were associated with dyslipidemia and lipid traits among Chinese children. In addition, we found significant gene-by-sex interactions. Our findings provided evidence supporting the role of FAM3C gene in regulating lipid metabolism in humans.
Impact
-
FAM3C genetic variants were associated with dyslipidemia and lipid traits among Chinese children. In addition, we found significant gene-by-sex interactions.
-
FAM3C/rs7776725 was associated with dyslipidemia and lipid traits only in girls.
-
Our findings provided evidence supporting the role of FAM3C gene in regulating lipid metabolism in humans.
Similar content being viewed by others
Introduction
Family with sequence similarity 3 member C (FAM3C), previously known as interleukin-like epithelial to mesenchymal transition inducer (ILEI), is a cytokine-like protein essential for epithelial to mesenchymal transition, tumor formation and metastasis in normal epithelial cells.1 Researches have demonstrated that FAM3C gene was involved in embryonic development, and the progression of pancreatic cancer.2,3 Furthermore, FAM3C was overexpressed and/or altered in intracellular localization in various human tumors, rendering it a promising molecular marker for the diagnosis and prognosis of some cancers, such as colon and breast cancer.1,2
Recently, Chen et al.4 performed functional studies and revealed a novel function of FAM3C in glucose and lipid metabolism in hepatocytes. Chen et al.4 identified that FAM3C suppressed the mTOR-SREBP1-FAS lipogenic pathway in hepatocytes in vitro and in vivo, which contributed to the beneficial effects of FAM3C on steatosis.4,5 They considered FAM3C as a novel therapeutical target for metabolic diseases, such as nonalcoholic fatty liver disease (NAFLD).4,5 However, whether FAM3C genetic variants are associated lipid traits among humans remain largely unknown. Moreover, previous genome-wide association studies and subsequent replication studies have found that genetic variants near FAM3C gene were associated with bone mineral density (BMD) and fracture in children,6,7,8 which is correlated with childhood overweight and obesity.9,10,11,12 Therefore, we hypothesized that FAM3C genetic variants would also be associated with overweight/obesity and lipid traits.
In the present study, we aimed to analyze the associations of FAM3C genetic variants with overweight/obesity and lipid traits among Chinese children; and assess whether the genetic associations differed in boys and girls.
Methods
Participants
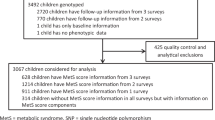
The current study subjects were from four independent studies conducted in Beijing, China, with a total sample size of 3305 children (1029 obese children, 1014 overweight children and 1262 normal-weight children). The study on adolescent lipids, insulin resistance and candidate genes (ALIR) and the Comprehensive Prevention project for Overweight and Obese Adolescents (CPOOA) were both case−control studies with similar subject recruitments and data collection methods, involving 937 (386 obese children, 400 overweight children and 151 normal-weight children) and 1093 children (319 obese children, 318 overweight children and 456 normal-weight children), respectively.13,14 The other two studies were school-based interventions on childhood and adolescent obesity. The School-Based Comprehensive Intervention on Childhood Obesity (SCICO) was performed in eight elementary schools of the Haidian District of Beijing, including 153 obese children, 151 overweight children and 1588 normal-weight children.15 The baseline survey data of all the overweight and obese children, as well as 152 normal-weight children randomly selected from the normal-weight group, were analyzed in the present study. The School-based Physical Activity Intervention on Childhood Obesity (SPAICO) was conducted in two primary schools and two middle schools in Changping District of Beijing.16 The cross-sectional baseline survey data of all children with blood samples in the SPAICO were analyzed in the current study, involving 171 obese children, 145 overweight children and 503 normal-weight children. Detailed descriptions of the four studies have been previously published.13,14,15,16 Subjects with any vital organs diseases and obesity-related comorbidities, including heart disease, liver diseases, pulmonary diseases, kidney disease and diabetes, were excluded, as well as those who were underweight, according to the Chinese national screening criteria for malnutrition of school-age children and adolescents.17
Written informed consents were obtained from all children and their parents. The ALIR, CPOOA and SPAICO were approved by the Ethic Committee of Peking University Health Science Center, and the SCICO was approved by the Ethic Committee of Chinese Center for Disease Control and Prevention. All studies abided by the Declaration of Helsinki principles.
Anthropometric measurements
Anthropometric measurements including height and weight in the four studies were performed according to the same standard protocols.13,14,15,16 We calculated BMI (kg/m2) using weight (in kilograms) divided by the square of height (in meters). Sex-and age-specific BMI standard deviation score (BMI-SDS) was also calculated, according to the growth reference data of the World Health Organization for children aged 5–19 years.18 The body fat percentage was measured using bioelectrical impedance analyzer except in ALIR study (CPOOA: Genuis-220, Jawon, Korea; SCICO and SPAICO: DF50, ImpediMed, Australia).
In the current study, obesity was defined as over 95th percentile of age- and sex-specific BMI and overweight was defined as between 85th and 95th percentile of age- and sex-specific BMI for children aged 7–18 years. The cut-offs were determined in a representative Chinese population.19 For children aged 6 years, the Chinese national screening criteria for overweight and obesity among school-age children and adolescents were applied.20
Biochemical assessment
Laboratory assays were performed on blood samples collected from all children after an overnight fast. Biochemical autoanalyzer (ALIR and CPOOA: Hitachi7060, Tokyo, Japan; SCICO and SPAICO: Olympus AU400, Tokyo, Japan) was used to determine total cholesterol, triglyceride, low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C). Dyslipidemia was defined according to the latest report of National Cholesterol Education Program (NCEP) Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents,21 as having one or more of the following: (1) an elevated TC level (TC ≥ 5.18 mmol/L); (2) an elevated TG level (children aged 7–9 years old: TG ≥ 1.13 mmol/L; children aged 10–19 years old: TG ≥ 1.47 mmol/L); (3) an elevated LDL-C level (LDL-C ≥ 3.37 mmol/L); (4) or a low HDL-C level (HDL-C ≤ 1.04 mmol/L).
SNPs selection and genotyping
Given the sample size, in order to reach over 80% power to detect an assumed effect size of 1.3, the allele frequency should be equal to or greater than 0.10. Therefore, only 21 intronic SNPs with minor allele frequency (MAF) ≥ 0.10 in the Han Chinese population from 1000 genomes (http://phase3browser.1000genomes.org/index.html) were left, representing by three tag SNPs. We only needed to select three SNPs, because the other SNPs were in strong linkage disequilibrium (LD) (r2 > 0.80) with these three tag SNPs. However, for one tag SNP, multiplex SNP assays design failed and no proxy SNP was available. Therefore, we finally included two intronic SNPs: rs7776725 (chromosome and position (based on NCBI build 38 (NCBI, Bethesda, MD)): 7:121393067) and rs7793554 (7:121393137) within the FAM3C gene (r2 = 0.61), representing >95% SNPs with MAF ≥ 0.10. Both rs7776725 and rs7793554 were previously reported to be associated with BMD and fracture.6,7,8 We used Haploview to estimate LD between the SNPs.22
Genomic DNA of all children was extracted from the blood leukocytes using the phenol-chloroform extraction method (ALIR, CPOOA and SCICO) and salt extraction method (SPAICO). Genotyping of rs7776725 and rs7793554 was conducted using the MassARRAY System (Agena Bioscience Inc., San Diego, CA, USA). The detail genotyping process had been described elsewhere.23,24 The call rates for rs7776725 and rs7793554 were 99.8% and 99.0%, respectively. For quality control, we performed genotyping on 5% randomly selected duplicated samples with 100% concordance.
Statistical analyses
Data were expressed as the mean and standard deviation (SD) for continuous variables and n (%) for categorical variables. Total cholesterol, triglyceride, LDL-C and HDL-C were natural log-transformed before analysis. The differences in general characteristics among normal-weight, overweight and obese children were examined with one-way analysis of variance (ANOVA) (continuous variables) or χ2 test (categorical variables). The Hardy−Weinberg equilibrium (HWE) of the genotype data in the normal-weight children were examined by a χ2 test.
The primary outcomes were overweight/obesity and dyslipidemia, and the secondly outcomes were anthropometric and lipid quantitative traits. We tested additive, dominant and recessive genetic models for the primary outcomes, and found that the additive model could best explain the effect of rs7776725 and rs7793554 based on the Akaike information criterion and the Bayesian information criterion. Therefore, the associations of FAM3C genetic variants with overweight/obesity and dyslipidemia were analyzed using logistic regressions, and their associations with anthropometric and lipid quantitative traits were analyzed using general linear regressions, assuming additive model adjusting for study groups, sex, age, age squared and BMI (except for overweight/obesity, BMI, and BMI-SDS). In addition, we performed sex-specific analyses and tested the gene-by-sex interaction effects. Since the participants were recruited from four independent studies, we also performed sensitivity analyses using meta-analysis with the inverse variance method.15,25
A two-sided P value of 0.05 was considered nominally significant. P < 0.003 (i.e. 0.05/[2SNPs × (2 categorical variables + 7 continuous variables)] = 0.05/18 = 0.003) was considered statistically significant after multiple testing using Bonferroni correction. We used Quanto 1.2.4 (University of Southern California, Los Angeles, CA, USA; http://biostats.usc.edu/cgi-bin/DownloadQuanto.pl) to calculate power and estimate the detectable effect sizes. All statistical analyses were performed with SPSS 24.0 (IBM Corp., Armonk, NY, USA). Meta-analyses were conducted with Revman 5.3.
Results
Table 1 presents the characteristics of the 3305 children in the normal-weight and overweight/obese groups. There were significant differences in sex and age between the two groups (P < 0.001). Compared with normal-weight children, overweight/obese children had higher BMI, body fat percentage, total cholesterol, triglyceride, LDL-C, and lower HDL-C (all P < 0.001).
The two SNPs (rs7776725 and rs7793554, located in the intron of the FAM3C gene) were in moderate LD (r2 = 0.61) in our study, and the minor allele frequencies were 0.112 and 0.168, respectively (Supplementary Table 1), similar to those reported in Han Chinese from 1000 genomes (http://phase3browser.1000genomes.org/index.html, 0.102 and 0.170 respectively). Among normal-weight children, the genotype frequencies of the two SNPs were both in HWE (P > 0.05, Supplementary Table 1).
Logistic regression analyses were performed to examine the associations of rs7776725 and rs7793554 with overweight/obesity and dyslipidemia. Both rs7776725 and rs7793554 were nominally associated with the risk of dyslipidemia (OR (95% CI) = 1.402 (1.116, 1.761), P = 0.004 and OR (95% CI) = 1.291 (1.069, 1.560), P = 0.008, respectively, Table 2), after adjustment for study groups, sex, age, age squared and BMI. However, these associations were no longer statistically significant after multiple correction.
We further analyzed the associations of the two FAM3C genetic variants with various anthropometric and lipid quantitative traits. As shown in Table 3, rs7776725 was nominally associated with triglyceride (β (SE) = 0.036 (0.015), P = 0.014) and HDL-C (β (SE) = −0.019 (0.008), P = 0.015), while rs7793554 was significantly associated with triglyceride (β (SE) = 0.040 (0.013), P = 0.001) and HDL-C (β (SE) = −0.019 (0.006), P = 0.003) after multiple correction. No significant associations were observed between the two FAM3C genetic variants and other anthropometric and lipid traits (all P > 0.05).
Sex-specific analysis
Subsequently, we performed sex-specific analysis. As presented in Fig. 1, rs7776725 was significantly associated with higher risk of dyslipidemia (OR (95% CI) = 2.119 (1.456, 3.083), P = 8.78 × 10–5) only in girls, and the interaction between rs7776725 and sex was nominally significant on the risk of dyslipidemia (Pfor interaction = 0.004). Similarly, rs7793554 was nominally associated with dyslipidemia only in girls (OR (95% CI) = 1.495 (1.108, 2.019), P = 0.009). However, the interaction between rs7793554 and sex was not significant (Pfor interaction = 0.183).
Logistic regressions were performed to explore the associations of rs7776725 (a) and rs7793554 (b) within the FAM3C gene with overweight/obesity and dyslipidemia in boys and girls, separately. Gene-by-sex interactions were also tested, assuming additive model and adjusting for study groups, age, age square and BMI (except for overweight/obesity). CI confidence interval, OR odds ratios.
Besides, we also found nominally significant interactions between rs7776725 and sex on total cholesterol and LDL-C (Pfor interaction = 0.012 and 0.008, respectively, Fig. 2). Girls with T-allele of rs7776725 were associated with higher total cholesterol (β = 0.030, P = 0.002) and LDL-C (β = 0.050, P = 0.001). Meanwhile, the interactions between rs7793554 and sex on total cholesterol and LDL-C were close to nominally significant level (Pfor interaction = 0.053 and 0.084, respectively, Supplementary Fig. 1). No significant gene-by-sex interactions were observed between the two FAM3C genetic variants and sex on other blood lipids, and anthropometric traits (all P > 0.05).
Linear regressions were performed to explore the associations of rs7776725 with total cholesterol (a), triglyceride (b), LDL-C (c), and HDL-C (d) in boys and girls, separately. Gene-by-sex interactions were also tested, assuming additive model and adjusting for study groups, age, age square and BMI. HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, TC total cholesterol, TG, triglyceride.
Sensitivity analyses
We also performed sensitivity analyses to pool the results of the four studies using meta-analysis. The results of sensitivity analyses were consistent with the primary results (Supplementary Tables 2, 3).
Discussion
In this association study among 3305 Chinese children, we examined the associations of FAM3C genetic variants with anthropometric and lipid traits. We found significant associations of rs7776725 and rs7793554 with dyslipidemia and lipid traits. In addition, we observed significant interactions between rs7776725 and sex on dyslipidemia, and sex-stratified analyses showed that it was significantly associated with dyslipidemia only in girls. The variant also showed significant interactions with sex on total cholesterol and LDL-C, with T-allele of rs7776725 associated with higher total cholesterol and LDL-C only in girls.
FAM3C is a member of the family with sequence similarity 3 family and expressed in almost all tissues.26 It encodes a secreted protein with a GG domain, which is a cytokine involved in epithelial to mesenchymal transition, tumor formation and metastasis in normal epithelial cells.1 In 2010, Katahira et al.27 reported its function in retinal laminar formation processes. Besides, previous studies also indicated that FAM3C gene could play an important role in bone metabolism.7,8,28,29,30,31 Both rs7776725 and rs7793554 are located in the first intron of the FAM3C gene, and the variant rs7793554 was suggested to be an intronic enhancer. The A to G change of rs7793554 will delete a binding site of the transcription factor CRE-BP (cyclic AMP response element (CRE)- binding protein (BP)).31
We found that FAM3C genetic variants (rs7776725 and rs7793554) were significantly associated with dyslipidemia and lipid profiles in children. Chen et al.4 firstly revealed that FAM3C played an important role in regulating lipid metabolism in hepatocytes in mice. Chen et al.4 performed functional studies and found that FAM3C overexpression reduced phosphorylated mechanistic target of rapamycin (mTOR) complex 1 (pmTORC1), SREBP-1c, and fatty acid synthase (FAS) protein levels in hepatic cells. Our findings provided supportive evidence for the function of FAM3C gene in lipid metabolism in humans. Moreover, we analyzed their associations with four quantitative lipid traits, including total cholesterol, triglyceride, LDL-C and HDL-C. We found that FAM3C rs7776725 T-allele and rs7793554 A-allele were associated with higher triglyceride and lower HDL-C in a total sample size of 3305 children. Although the associations of FAM3C genetic variants with triglyceride and HDL-C attenuated or abolished after stratified by sex, the effect sizes pointed into the same direction in boys and girls as in the total sample. This failed replication after stratification by sex might be a problem of reduced statistic power due to smaller sample size in each subgroup (power 34−65%). However, neither of the SNPs had been previously reported to be associated with lipids traits by large-scale genome-wide association studies.32,33,34,35 The reason might be that this is a children-specific effect, because the previous studies were performed in adult populations. The genetics of these lipids traits in children remained largely unknown. Therefore, our study could help to provide more information about the genes associated with lipid metabolism, especially in Chinese children. More validation studies with larger sample sizes are needed.
In addition, we found the genetic associations showed significant sex differences. We noted that rs7776725 was significantly associated with dyslipidemia, total cholesterol and LDL-C only in girls (power > 90%). Similar genetic associations were also observed in rs7793554, although the gene-by-sex interactions were only marginally significant. However, the underlying molecular mechanism remains unclear, though it was suggested that sexual dimorphism might be at least partly explained by the differences in levels of sex-hormones.36 Other researchers provided a novel insight into sex-specific differences of cell regulatory processes.37 More researches are required before a clear understanding of biochemical mechanisms underlying sexual dimorphism in the future.
Although some evidences have shown that genetic variants near FAM3C gene were associated with BMD and fracture in children,6,7,8 which were correlated with childhood overweight and obesity.9,10,11,12 In the present study, we did not find rs7776725 and rs7793554 were associated with the risk of overweight/obesity. And no previous studies have reported the associations between FAM3C genetic variants and childhood overweight and obesity. More studies are awaited to confirm our findings.
Strengths and limitations
To the best of our knowledge, this is the first study to identify significant associations of FAM3C genetic variants with dyslipidemia and lipid traits among Chinese children. Our study provided supportive evidence for the function of FAM3C gene in regulating lipid metabolism.
There were also several limitations in our study. Firstly, the heterogeneity among the four independent studies should be considered. However, we have adjusted for the study groups in the regression models. And our previous studies also showed that there was little heterogeneity among the four independent studies.15,25 Secondly, there might be population stratification. However, we thought the population stratification effect was likely to be small because of the following two reasons. On the one hand, the heterogeneity among the four independent studies was generally small (I2 < 30%, P for heterogeneity < 0.05). And it is reasonable to assume that all participants were selected from a random mating population. Therefore, in theory, we thought participants in our study should have high homogeneity in genetic background. On the other hand, almost all children in our study were Han Chinese (about 94%); thus the systematic difference in allele frequencies between subpopulations would be small. Thirdly, as no previous studies have reported the association between the FAM3C genetic variants and overweight/obesity or dyslipidemia, we could only estimate the statistic power. Based on the sample size and effect estimates in the current study, the power to detect associations of FAM3C genetic variants with dyslipidemia and gene-by-sex interaction were >80%. But the power for overweight/obesity was <60%, which need further validation studies with larger sample size.
Conclusions
In conclusion, we identified for the first time that FAM3C genetic variants (rs7776725 and rs7793554) were associated with dyslipidemia and lipid traits among Chinese children. In addition, we found significant interactions between rs7776725 and sex on dyslipidemia, total cholesterol and LDL-C traits. Our findings provided evidence supporting the role of FAM3C gene in regulating lipid metabolism in humans. More studies are awaited to confirm our findings in the future.
References
Waerner, T. et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–239 (2006).
Yang, J. & Guan, Y. Family with sequence similarity 3 gene family and nonalcoholic fatty liver disease. J. Gastroenterol. Hepatol. 28(Suppl 1), 105–111 (2013).
Gronborg, M. et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol. Cell Proteom. 5, 157–171 (2006).
Chen, Z. et al. Hepatic activation of the FAM3C-HSF1-CaM pathway attenuates hyperglycemia of obese diabetic mice. Diabetes 66, 1185–1197 (2017).
Zhang, X. et al. FAM3 gene family: a promising therapeutical target for NAFLD and type 2 diabetes. Metabolism 81, 71–82 (2018).
Chesi, A. et al. A trans-ethnic genome-wide association study identifies gender-specific loci influencing pediatric aBMD and BMC at the distal radius. Hum. Mol. Genet. 24, 5053–5059 (2015).
Kemp, J. P. et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 10, e1004423 (2014).
Medina-Gomez, C. et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 8, e1002718 (2012).
van Leeuwen, J., Koes, B. W., Paulis, W. D. & van Middelkoop, M. Differences in bone mineral density between normal-weight children and children with overweight and obesity: a systematic review and meta-analysis. Obes. Rev. 18, 526–546 (2017).
Ellis, K. J., Shypailo, R. J., Wong, W. W. & Abrams, S. A. Bone mineral mass in overweight and obese children: diminished or enhanced? Acta Diabetol. 40(Suppl 1), S274–S277 (2003).
Gracia-Marco, L. et al. Adiposity and bone health in Spanish adolescents. The HELENA study. Osteoporos. Int. 23, 937–947 (2012).
Klein, K. O. et al. Effect of obesity on estradiol level, and its relationship to leptin, bone maturation, and bone mineral density in children. J. Clin. Endocrinol. Metab. 83, 3469–3475 (1998).
Wang, H.-J. et al. Association of the common genetic variant upstream of INSIG2 gene with obesity related phenotypes in Chinese children and adolescents. Biomed. Environ. Sci. 21, 528–536 (2008).
Wang, D. et al. Association of the MC4R V103I polymorphism with obesity: a Chinese case-control study and meta-analysis in 55,195 individuals. Obesity (Silver Spring) 18, 573–579 (2010).
Song, Q.-Y. et al. Association study of three gene polymorphisms recently identified by a genome-wide association study with obesity-related phenotypes in Chinese children. Obes. Facts 10, 179–190 (2017).
Li, X.-H. et al. Effectiveness of a school-based physical activity intervention on obesity in school children: a nonrandomized controlled trial. BMC Public Health 14, 1282 (2014).
National Health and Family Planning Commission of PRC. Screening standard for malnutrition of school-age children and adolescents. WS/T 456-2014 (NHFPC, China, 2014).
World Health Organization. Growth reference data for 5−19 years. (2007). http://www.who.int/growthref/en/.
Ji, C.-Y. Report on childhood obesity in China (1)–body mass index reference for screening overweight and obesity in Chinese school-age children. Biomed. Environ. Sci. 18, 390–400 (2005).
National Health and Family Planning Commission of PRC. Screening for overweight and obesity among school-age children and adolescents. WS/T 586-2018 (NHFPC, China, 2018).
Expert Panel on Integrated Guidelines for Cardiovascular Health and Risk Reduction in Children and Adolescents. Summary report. Pediatrics 128(Suppl S), 213–256 (2011).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Wang, H.-J. et al. Association of common variants identified by recent genome-wide association studies with obesity in Chinese children: a case-control study. BMC Med. Genet. 17, 7 (2016).
Meng, X.-R. et al. Association study of childhood obesity with eight genetic variants recently identified by genome-wide association studies. Pediatr. Res. 76, 310–315 (2014).
Song, Q. et al. Physical activity attenuates the association between the IRS1 genotype and childhood obesity in Chinese children. Nutr. Metab. Cardiovasc. Dis. 29, 793–801 (2019).
Zhu, Y. et al. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics 80, 144–150 (2002).
Katahira, T., Nakagiri, S., Terada, K. & Furukawa, T. Secreted factor FAM3C (ILEI) is involved in retinal laminar formation. Biochem. Biophys. Res. Commun. 392, 301–306 (2010).
Estrada, K. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501 (2012).
Cho, Y. S. et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 41, 527–534 (2009).
Zheng, H.-F. et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 8, e1002745 (2012).
Zhang, L.-S. et al. A follow-up association study of two genetic variants for bone mineral density variation in Caucasians. Osteoporos. Int. 23, 1867–1875 (2012).
Hoffmann, T. J. et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 50, 401–413 (2018).
Sandhu, M. S. et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet 371, 483–491 (2008).
Bentley, A. R. et al. Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat. Genet. 51, 636–648 (2019).
Spracklen, C. N. et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 26, 1770–1784 (2017).
Wisniewski, A. B. & Chernausek, S. D. Gender in childhood obesity: family environment, hormones, and genes. Gend. Med. 6(Suppl 1), 76–85 (2009).
Mittelstrass, K. et al. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 7, e1002215 (2011).
Acknowledgements
We thank all the children and adolescents and their parents for their participation. The study was supported by grants from the National Natural Science Foundation of China (grant recipient: H.W., grant number: 81573170). Q.S. is funded by the China Scholarship Council for her study in the United States (201806010383).
Author information
Authors and Affiliations
Contributions
Substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data: all authors. Drafting the article or revising it critically for important intellectual content: Q.S., J.S., L.Q., and H.W. Final approval of the version to be published: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
In the present study, written informed consents were obtained from all children and their parents.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Song, Q., Song, J., Li, C. et al. Genetic variants in the FAM3C gene are associated with lipid traits in Chinese children. Pediatr Res 89, 673–678 (2021). https://doi.org/10.1038/s41390-020-0897-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0897-3