Abstract
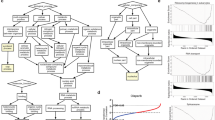
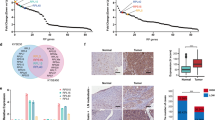
Topoisomerase II poisons are one of the most common class of chemotherapeutics used in cancer. We and others had shown that a subset of glioblastomas, the most malignant of all primary brain tumors in adults, is responsive to TOP2 poisons. To identify genes that confer susceptibility to this drug in gliomas, we performed a genome-scale CRISPR knockout screen with etoposide. Genes involved in protein synthesis and DNA damage were implicated in etoposide susceptibility. To define potential biomarkers for TOP2 poisons, CRISPR hits were overlapped with genes whose expression correlates with susceptibility to this drug across glioma cell lines, revealing ribosomal protein subunit RPS11, 16, and 18 as putative biomarkers for response to TOP2 poisons. Loss of RPS11 led to resistance to etoposide and doxorubicin and impaired the induction of proapoptotic gene APAF1 following treatment. The expression of these ribosomal subunits was also associated with susceptibility to TOP2 poisons across cell lines from gliomas and multiple other cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318:2306–16.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110.
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–37.
Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–63.
Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–50.
Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–37.
Mehta A, Awah CU, Sonabend AM. Topoisomerase II poisons for glioblastoma; existing challenges and opportunities to personalize therapy. Front Neurol. 2018;9:459.
Sonabend AM, Carminucci AS, Amendolara B, Bansal M, Leung R, Lei L, et al. Convection-enhanced delivery of etoposide is effective against murine proneural glioblastoma. Neuro Oncol. 2014;16:1210–9.
Kesari S, Schiff D, Doherty L, Gigas DC, Batchelor TT, Muzikansky A, et al. Phase II study of metronomic chemotherapy for recurrent malignant gliomas in adults. Neuro Oncol. 2007;9:354–63.
Reardon DA, Desjardins A, Vredenburgh JJ, Gururangan S, Sampson JH, Sathornsumetee S, et al. Metronomic chemotherapy with daily, oral etoposide plus bevacizumab for recurrent malignant glioma: a phase II study. Br J Cancer. 2009;101:1986–94.
Leonard A, Wolff JE. Etoposide improves survival in high-grade glioma: a meta-analysis. Anticancer Res. 2013;33:3307–15.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD. et al. Genome-scale CRISPR-Cas9 knockout transcriptional activation Screen. Nat Protoc. 2017;12:828–63.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87.
Pitz MW, Desai A, Grossman SA, Blakeley JO. Tissue concentration of systemically administered antineoplastic agents in human brain tumors. J Neurooncol. 2011;104:629–38.
Zucchetti M, Rossi C, Knerich R, Donelli MG, Butti G, Silvani V, et al. Concentrations of VP16 and VM26 in human brain tumors. Ann Oncol. 1991;2:63–6.
Stewart DJ, Richard MT, Hugenholtz H, Dennery JM, Belanger R, Gerin-Lajoie J, et al. Penetration of VP-16 (etoposide) into human intracerebral and extracerebral tumors. J Neurooncol. 1984;2:133–9.
Koschmann C, Calinescu AA, Nunez FJ, Mackay A, Fazal-Salom J, Thomas D, et al. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016;8:328ra28.
Michlits G, Hubmann M, Wu SH, Vainorius G, Budusan E, Zhuk S, et al. CRISPR-UMI: single-cell lineage tracing of pooled CRISPR-Cas9 screens. Nat Methods. 2017;14:1191–7.
Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4.
Wijdeven RH, Pang B, van der Zanden SY, Qiao X, Blomen V, Hoogstraat M, et al. Genome-wide identification and characterization of novel factors conferring resistance to topoisomerase II poisons in cancer. Cancer Res. 2015;75:4176–87.
Cancer Cell Line Encyclopedia, C. and C. Genomics of Drug Sensitivity in Cancer. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528:84–7.
Forbes SA, Beare D, Bindal N, Bamford S, Ward S, Cole CG, et al. COSMIC: high-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr Protoc Hum Genet. 2016;91:10111–37.
Uuskula-Reimand L, Hou H, Samavarchi-Tehrani P, Rudan MV, Liang M, Medina-Rivera A, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17:182.
Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–49.
Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–56.
Forester CM. et al. Revealing nascent proteomics in signaling pathways and cell differerentiation. Proc Natl Acad Sci USA. 2018;115:2353–8.
Sapio RT, Nezdyur AN, Krevetski M, Anikin L, Manna VJ, Minkovsky N, et al. Inhibition of post-transcriptional steps in ribosome biogenesis confers cytoprotection against chemotherapeutic agents in a p53-dependent manner. Sci Rep. 2017;7:9041.
Pop C, Salvesen GS. The nematode death machine in 3D. Cell. 2005;123:192–3.
Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22:269–75.
Zinkel SS, Hurov KE, Ong C, Abtahi FM, Gross A, Korsmeyer S. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–91.
Mjelle R, Hegre SA, Aas PA, Slupphaug G, Drabløs F, Saetrom P, et al. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair. 2015;30:53–67.
Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, et al. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–32.
Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–30.
Barnoud D, Pinçon C, Bruno B, Béné J, Gautier S, Lahoche A, et al. Acute kidney injury after high dose etoposide phosphate: a retrospective study in children receiving an allogeneic hematopoetic stem cell transplantation. Pediatr Blood Cancer. 2018;65:e27038.
Girling DJ. Comparison of oral etoposide and standard intravenous multidrug chemotherapy for small-cell lung cancer: a stopped multicentre randomised trial. Medical Research Council Lung Cancer Working Party. Lancet. 1996;348:563–6.
Franceschi E, Cavallo G, Scopece L, Paioli A, Pession A, Magrini E, et al. Phase II trial of carboplatin and etoposide for patients with recurrent high-grade glioma. Br J Cancer. 2004;91:1038–44.
Krisp, C, Parker R, Pascovici D, Hayward NK, Wilmott JS, Thompson JF, et al. Proteomic phenotyping of metastatic melanoma reveals putative signatures of MEK inhibitor response and prognosis. Br J Cancer. 2018.
Moreno P, Jim‚nez-Jim‚nez C, Garrido-Rodr¡guez M, Calder¢n-Santiago M, Molina S, Lara-Chica M. Metabolomic profiling of human lung tumor tissues: nucleotide metabolism as a candidate for therapeutic interventions and biomarkers. Mol Oncol. 2018;12:1778–96.
Tsai CH, Chen YT, Chang YH, Hsueh C, Liu CY, Chang YS, et al. Systematic verification of bladder cancer-associated tissue protein biomarker candidates in clinical urine specimens. Oncotarget. 2018;9:30731–47.
Idbaih A, Canney M, Belin L, Desseaux C, Vignot A, Bouchoux G, et al. Safety and feasibility of repeated and transient blood-brain barrier disruption by pulsed ultrasound in patients with recurrent glioblastoma. Clin Cancer Res. 2019;25:3793–801.
Carpentier A, Canney M, Vignot A, Reina V, Beccaria K, Horodyckid C, et al. Clinical trial of blood-brain barrier disruption by pulsed ultrasound. Sci Transl Med. 2016;8:343re2. https://doi.org/10.1126/scitranslmed.aaf6086.
Mainprize T, Lipsman N, Huang Y, Meng Y, Bethune A, Ironside S, et al. Blood-brain barrier opening in primary brain tumors with non-invasive MR-guided focused ultrasound: a clinical safety and feasibility study. Sci Rep. 2019;9:321 https://doi.org/10.1038/s41598-018-36340-0.
Alli S, Figueiredo CA, Golbourn B, Sabha N, Wu MY, Bondoc A, et al. Brainstem blood brain barrier disruption using focused ultrasound: a demonstration of feasibility and enhanced doxorubicin delivery. J Control Release. 2018;281:29–41. https://doi.org/10.1016/j.jconrel.2018.05.005.
Lin YL, Wu MT, Yang FY. Pharmacokinetics of doxorubicin in glioblastoma multiforme following ultrasound-Induced blood-brain barrier disruption as determined by microdialysis. J Pharm Biomed Anal. 2018;149:482–7. https://doi.org/10.1016/j.jpba.2017.11.047.
Sun T, Zhang Y, Power C, Alexander PM, Sutton JT, Aryal M, et al. Closed-loop control of targeted ultrasound drug delivery across the blood-brain/tumor barriers in a rat glioma model. Proc Natl Acad Sci USA. 2017;114:E10281–E10290. https://doi.org/10.1073/pnas.1713328114.
Sonabend AM, Stupp R. Overcoming the blood brain barrier with an implantable ultrasound device. Clin Cancer Res. 2019;25:3750–2. https://doi.org/10.1158/1078-0432.
Winter. J Schwering M, Pelz O, Rauscher B, Zhan T, Heigwer F, et al. CRISPRAnalyzeR: interactive analysis, annotation and documentation of pooled CRISPR screens. 2017. https://doi.org/10.1101/109967.
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3.
Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3.
Miller AD, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–902.
Acknowledgements
This work was funded by 5DP5OD021356-05 (AMS), P50CA221747 SPORE for Translational Approaches to Brain Cancer (AMS), and Developmental funds from The Robert H. Lurie NCI Cancer Center Support Grant #P30CA060553 (AMS), and generous philanthropic support from Dan and Sharon Moceri. We thank Dr. Ichiro Nakano (University of Alabama), Dr. Charles David James (Northwestern University), and Dr. Shi-Yuan Cheng (Northwestern University) for the kind gift of the GBM xenografts. We thank Dr. Peng Zhang (Northwestern University) for technical support with culture of GBM PDX. We thank Synthego, CA, USA, for the gifts of RPS11 sgRNA guides. Immunofluorescence imaging work was performed at the Center for Advanced Microscopy/Nikon Imaging Center and RHLCCC - Flow Cytometry Core, Northwestern University supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. BG was supported by the NVKP 16-1-2016-0037 grant.
Author information
Authors and Affiliations
Contributions
AMS and CUA conceived the idea of this project. CUA, AM, and LC performed the genome-scale CRISPR screen. JW and CUA performed the screen analysis. MB and CUA performed the combinatorial analysis to predict biomarkers. CUA performed the single-gene editing and all validations. ML contributed to organizing and curating the large-scale data used in this work. CUA, LW, and ZD performed western blots. EF contributed in cell culture. DY and CUA performed the immunohistochemistry, EBG and CP transduced lentivirus with RPS11 overexpression vector. MZ and CUA performed the flow cytometry analysis. SK contributed to the CRISPR screen. IVB contributed reagents and corrected the manuscript. CUA and RM performed the DNA damage analysis on edited cells and GBM cell lines, respectively, and LC performed 53BP1 staining. BG, KBB, and DMS performed oversaw statistical analysis. AMS, PH, and AH supervised the project. CUA, MZ, and AMS prepared figures. CUA and AMS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare a conflict of interest as a patent has been filed based on the findings from this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Awah, C.U., Chen, L., Bansal, M. et al. Ribosomal protein S11 influences glioma response to TOP2 poisons. Oncogene 39, 5068–5081 (2020). https://doi.org/10.1038/s41388-020-1342-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1342-0
This article is cited by
-
A Four-Gene Panel for the Prediction of Prognosis and Immune Cell Enrichment in Gliomas
Molecular Biotechnology (2023)
-
Comprehensive proteomic analysis of exosome mimetic vesicles and exosomes derived from human umbilical cord mesenchymal stem cells
Stem Cell Research & Therapy (2022)