Abstract
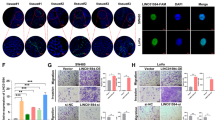
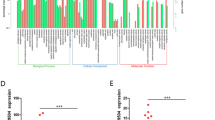
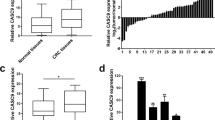
Long noncoding RNAs (lncRNAs) have been shown to play crucial roles in cancer long noncoding RNAs (lncRNAs) have been known to play crucial roles in cancer development and progression by regulating chromatin dynamics and gene expression. However, only a few lncRNAs with annotated functions in the progression of colorectal cancer (CRC) have been identified to date. In the present study, the expression of lncCMPK2 was upregulated in CRC tissues and positively correlated with clinical stages and lymphatic metastasis. The overexpression of lncCMPK2 promoted the proliferation and cell cycle transition of CRC cells. Conversely, the silencing of lncCMPK2 restricted cell proliferation both in vitro and in vivo. lncCMPK2 was localized to the nucleus of CRC cells, bound to far upstream element binding protein 3 (FUBP3), and guided FUBP3 to the far upstream element (FUSE) of the c-Myc gene to activate transcription. lncCMPK2 also stabilized FUBP3. These results provide novel insights into the functional mechanism of lncCMPK2 in CRC progression and highlight its potential as a biomarker of advanced CRC and therapeutic target.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data and material are available in Supplementary Data.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91.
Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357:55–60.
Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932–56.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9.
Liu J, Peng WX, Mo YY, Luo D. MALAT1-mediated tumorigenesis. Front Biosci (Landmark Ed). 2017;22:66–80.
Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505.
Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61.
Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–14.
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that Interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–20.e709.
Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48:194–207.
Malakar P, Stein I, Saragovi A, Winkler R, Stern-Ginossar N, Berger M, et al. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Res. 2019;79:2480–93.
Liu T, Han Z, Li H, Zhu Y, Sun Z, Zhu A. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17:118.
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–48.
Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61.
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2016;65:1494–504.
Han Q, Xu L, Lin W, Yao X, Jiang M, Zhou R, et al. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene. 2019;38:3019–32.
Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538–45.
Zheng P, Yin Z, Wu Y, Xu Y, Luo Y, Zhang TC. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun Signal. 2018;16:5.
Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9:e1003705.
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long non-coding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-Catenin pathway by scaffolding EZH2. Clin Cancer Res. 2018;24:684–95.
Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42:10668–80.
Mariotti B, Servaas NH, Rossato M, Tamassia N, Cassatella MA, Cossu M, et al. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front Immunol. 2019;10:100.
Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561:132–6.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19.
Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9.
Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–80.
Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S, et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer. 2008;8:369.
Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q, Chung MC. 2-D DIGE profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (FUBP) as novel candidates in liver carcinogenesis. Proteomics. 2008;8:5086–96.
Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271:31679–87.
Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35.
Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–64.
Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29.
Gonzalez V, Hurley LH. The c-MYC NHE III(1): function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–29.
Quinn LM. FUBP/KH domain proteins in transcription: back to the future. Transcription. 2017;8:185–92.
Huang H-I, Chang Y-Y, Lin J-Y, Kuo R-L, Liu H-P, Shih S-R, et al. Interactome analysis of the EV71 5′ untranslated region in differentiated neuronal cells SH-SY5Y and regulatory role of FBP3 in viral replication. Proteomics. 2016;16:2351–62.
Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell. 2017;171:1301–15.e1314.
Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–9.
Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang S, et al. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 2010;59:1226–35.
Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang F, et al. LIM and SH3 protein 1 induces TGFbeta-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin Cancer Res. 2014;20:5835–47.
Acknowledgements
We would like to thank Editage [www.editage.cn] and BioMed Proofreading@ LLC [www.biomedproofreading.com] for English language editing.
Funding
This work was supported by the National Natural Science Foundation of China (nos. 81572813, 81773082, 81702903, and 81872423), Guangdong Natural Science Foundation (2016A030313626, 2017A030310038, 2018B030311036), and Fork Ying Tung Education Foundation (161035).
Author information
Authors and Affiliations
Contributions
LZ led the study design and prepared the paper. Q-ZG and RZ carried out the experiments. YM and R-FD performed statistical analysis; LW and RL assisted in tissue sample collection. F-LD performed data analysis and interpretation. CL collected the data.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All experiments performed are endorsed by the Ethics Committee of Southern Medical University and complied with the Declaration of Helsinki. All animal experiments were carried out with the approval of the Southern Medical University Animal Care and Use Committee in accordance with the guidelines for the ethical treatment of animals. All animal experiments involved ethical and humane treatment under a license from the Guangdong Provincial Bureau of Science.
Informed consent
No informed consent was required because data were analyzed anonymously.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Gao, Q., Zhou, R., Meng, Y. et al. Long noncoding RNA CMPK2 promotes colorectal cancer progression by activating the FUBP3–c-Myc axis. Oncogene 39, 3926–3938 (2020). https://doi.org/10.1038/s41388-020-1266-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1266-8
This article is cited by
-
Decoding the regulatory landscape of lncRNAs as potential diagnostic and prognostic biomarkers for gastric and colorectal cancers
Clinical and Experimental Medicine (2024)
-
Therapeutic targeting of FUBP3 phase separation by GATA2-AS1 inhibits malate-aspartate shuttle and neuroblastoma progression via modulating SUZ12 activity
Oncogene (2023)
-
Identification of a key glioblastoma candidate gene, FUBP3, based on weighted gene co-expression network analysis
BMC Neurology (2022)
-
LCAT1 is an oncogenic LncRNA by stabilizing the IGF2BP2-CDC6 axis
Cell Death & Disease (2022)
-
Significant position of C-myc in colorectal cancer: a promising therapeutic target
Clinical and Translational Oncology (2022)