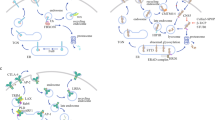
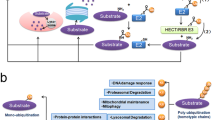
Abstract
Cancer immunotherapy using immune-checkpoint blockade has displayed promising clinical effects, but prevalent antibody-based inhibitors face multiple challenges such as low response rate, acquired resistance, and adverse effects. The intracellular expression of PD-1/PD-L1 in recycling endosomes and their active trafficking to membrane highlight the importance of depleting rather than interfering with checkpoint proteins. Preclinical investigations on the therapeutic effects of lead compounds that function by degrading immune checkpoint ligands and receptors have reported highly promising results. By harnessing the degradation capabilities of the lysosome, proteasome and autophagosomes, different small molecules and peptides potently induced degradation of checkpoint proteins and enhanced anti-tumor immunity. Both in vitro and in vivo experiments support the therapeutic efficacy of these molecules. Thus, targeted degradation through endo-lysosomal, autophagic, proteasomal, or endoplasmic reticulum-related pathways may provide promising strategies for tackling the challenges in cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Cha JH, Chan LC, Song MS, Hung MC. New approaches on cancer immunotherapy. Cold Spring Harbor perspectives in medicine. 2019;10:a036863.
Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med. 2019;25:656–66.
Hakenberg OW. Nivolumab for the treatment of bladder cancer. Expert Opin Biol Ther. 2017;17:1309–15.
Greillier L, Tomasini P, Barlesi F. The clinical utility of tumor mutational burden in non-small cell lung cancer. Transl Lung Cancer Res. 2018;7:639–46.
Esfahani K, Buhlaiga N, Thebault P, Lapointe R, Johnson NA, Miller WH Jr. Alemtuzumab for Immune-Related Myocarditis Due to PD-1 Therapy. N Engl J Med. 2019;380:2375–6.
Wang Y, Zhou S, Yang F, Qi X, Wang X, Guan X, et al. Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials: a systematic review and meta-analysis. JAMA Oncol. 2019;5:1008–19.
Liu Y, Wang H, Deng J, Sun C, He Y, Zhou C. Toxicity of tumor immune checkpoint inhibitors-more attention should be paid. Transl Lung Cancer Res. 2019;8:1125–33.
Hsu JM, Li CW, Lai YJ, Hung MC. Posttranslational Modifications of PD-L1 and Their Applications in Cancer Therapy. Cancer Res. 2018;78:6349–53.
Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell. 2019;76:359–70.
Chen S, Song Z, Zhang A. Small-molecule immuno-oncology therapy: advances, challenges and new directions. Curr Top Med Chem. 2019;19:180–5.
Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–5.
Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, et al. Melanoma Cell-Intrinsic PD-1 Receptor Functions Promote Tumor Growth. Cell. 2015;162:1242–56.
Li H, Li X, Liu S, Guo L, Zhang B, Zhang J, et al. Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology. 2017;66:1920–33.
Pu N, Gao S, Yin H, Li JA, Wu W, Fang Y, et al. Cell-intrinsic PD-1 promotes proliferation in pancreatic cancer by targeting CYR61/CTGF via the hippo pathway. Cancer Lett. 2019;460:42–53.
Theivanthiran B, Evans KS, DeVito NC, Plebanek MP, Sturdivant M, Wachsmuth LP. et al. A tumor-intrinsic PD-L1-NLRP3 inflammasome signaling pathway drives resistance to anti-PD-1 immunotherapy. J Clin Investig. 2020;130:2570–86.
Yao H, Wang H, Li C, Fang JY, Xu J. Cancer Cell-Intrinsic PD-1 and Implications in Combinatorial Immunotherapy. Front Immunol. 2018;9:1774.
Brosseau JP. Regulations on Messenger RNA: Wires and Nodes. Adv Exp Med Biol. 2020;1248:251–63.
Wang Y, Deng S, Xu J. Proteasomal and lysosomal degradation for specific and durable suppression of immunotherapeutic targets. Cancer Biol Med. 2020;17:583–98.
Liang L, Wang H, Shi H, Li Z, Yao H, Bu Z, et al. A Designed Peptide Targets Two Types of Modifications of p53 with Anti-cancer Activity. Cell Chem Biol. 2018;25:761–.e765.
Sun X, Gao H, Yang Y, He M, Wu Y, Song Y, et al. PROTACs: great opportunities for academia and industry. Signal Transduct Target Ther. 2019;4:64.
Dong P, Xiong Y, Yue J, Hanley SJB, Watari H. Tumor-Intrinsic PD-L1 Signaling in Cancer Initiation, Development and Treatment: Beyond Immune Evasion. Front Oncol. 2018;8:386.
Wang H, Yao H, Li C, Shi H, Lan J, Li Z, et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat Chem Biol. 2019;15:42–50.
Gao H, Yang Z, Zhang S, Cao S, Shen S, Pang Z, et al. Ligand modified nanoparticles increases cell uptake, alters endocytosis and elevates glioma distribution and internalization. Sci Rep. 2013;3:2534.
Kiran S, Dwivedi P, Khatik R, Hameed S, Dwivedi M, Huang F, et al. Synthesis of a functionalized dipeptide for targeted delivery and pH-sensitive release of chemotherapeutics. Chem Commun (Camb). 2019;56:285–8.
Dua P,SS, Kim S, Lee DK. ALPPL2 Aptamer-Mediated Targeted Delivery of 5-Fluoro-2’-Deoxyuridine to Pancreatic Cancer. Nucleic Acid Ther. 2015;25:180–7.
Lyle C, Richards S, Yasuda K, Napoleon MA, Walker J, Arinze N, et al. c-Cbl targets PD-1 in immune cells for proteasomal degradation and modulates colorectal tumor growth. Sci Rep. 2019;9:20257.
Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z, et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. 2018;564:130–5.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95.
Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7:4004–11.
Yao H, Xu J. Regulation of Cancer Immune Checkpoint: Mono- and Poly-Ubiquitination: Tags for Fate. Adv Exp Med Biol. 2020;1248:295–324.
Delport A, Hewer R. Inducing the Degradation of Disease-Related Proteins Using Heterobifunctional Molecules. Molecules. 2019;24:3272.
Wang Y, Jiang X, Feng F, Liu W, Sun H. Degradation of proteins by PROTACs and other strategies. Acta Pharm Sin B. 2020;10:207–38.
Neklesa T, Snyder LB, Willard RR, Vitale N, Pizzano J, Gordon DA, et al. ARV-110: An oral androgen receptor PROTAC degrader for prostate cancer. J Clin Oncol. 2019;37:259–259.
Dvela-Levitt M, Kost-Alimova M, Emani M, Kohnert E, Thompson R, Sidhom EH, et al. Small Molecule Targets TMED9 and Promotes Lysosomal Degradation to Reverse Proteinopathy. Cell. 2019;178:521–35 e523.
Cha JH, Yang WH, Xia W, Wei Y, Chan LC, Lim SO, et al. Metformin Promotes Antitumor Immunity via Endoplasmic-Reticulum-Associated Degradation of PD-L1. Mol Cell. 2018;71:606–20 e607.
Verdura S, Cuyàs E, Cortada E, Brunet J, Lopez-Bonet E, Martin-Castillo B, et al. Resveratrol targets PD-L1 glycosylation and dimerization to enhance antitumor T-cell immunity. Aging (Albany NY). 2020;12:8–34.
Wei Z, Zhang M, Li C, Huang W, Fan Y, Guo J, et al. Specific TBC Domain-Containing Proteins Control the ER-Golgi-Plasma Membrane Trafficking of GPCRs. Cell Rep. 2019;28:554–66 e554.
Gomez-Navarro N, Miller E. Protein sorting at the ER-Golgi interface. J Cell Biol. 2016;215:769–78.
Wang H, Han X, Xu J. Lysosome as the Black Hole for Checkpoint Molecules. Adv Exp Med Biol. 2020;1248:325–46.
Deng S, Zhou X, Xu J. Checkpoints under traffic control: from and to organelles. Adv Exp Med Biol. 2020;1248:431–53.
Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8:1239–42.
Riggs NL, Craig HM, Pandori MW, Guatelli JC. The dileucine-based sorting motif in HIV-1 Nef is not required for down-regulation of class I MHC. Virology. 1999;258:203–7.
Steven B, Kayvon P, Simon W, Nicholas R, Carolyn B. Lysosome Targeting Chimeras (LYTACs) for the degradation of secreted and membrane proteins. 2019;584:291–7.
Endicott SJ, Boynton DN, Jr., Beckmann LJ, Miller RA. Long-lived mice with reduced growth hormone signaling have a constitutive upregulation of hepatic chaperone-mediated autophagy. Autophagy. 2020: 1–14.
Fan X, Jin WY, Lu J, Wang J, Wang YT. Rapid and reversible knockdown of endogenous proteins by peptide-directed lysosomal degradation. Nat Neurosci. 2014;17:471–80.
Valdor R, Garcia-Bernal D, Riquelme D, Martinez CM, Moraleda JM, Cuervo AM, et al. Glioblastoma ablates pericytes antitumor immune function through aberrant up-regulation of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2019;116:20655–65.
Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96.
Wang X, Wu WKK, Gao J, Li Z, Dong B, Lin X, et al. Autophagy inhibition enhances PD-L1 expression in gastric cancer. J Exp Clin Cancer Res. 2019;38:140.
Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng. 2019;3:306–17.
Yao H, Li C, Brosseau J-P, Wang H, Lu H, Fang C et al. Palmitoylation is critically required for cancer intrinsic PD-1 expression and functions. bioRxiv. 2019: 625483.
Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Disco. 2019;18:41–58.
Baek K, Schulman BA. Molecular glue concept solidifies. Nat Chem Biol. 2020;16:2–3.
Chamberlain PP, Hamann LG. Development of targeted protein degradation therapeutics. Nat Chem Biol. 2019;15:937–44.
Yang Q, Peng L, Wu Y, Li Y, Wang L, Luo JH, et al. Endocytic adaptor protein HIP1R controls intracellular trafficking of epidermal growth factor receptor in neuronal dendritic development. Front Mol Neurosci. 2018;11:447.
Gao H, Sun X, Rao Y. PROTAC technology: opportunities and challenges. ACS Med Chem Lett. 2020;11:237–40.
Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Disco. 2017;16:19–34.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No: 82030104, 81874050, 81572326), Basic Research Projects of Shanghai Science and Technology Innovation Action Plan (20JC1410700); National Key R & D Program of China (2016YFC0906002, 2016YFC0906002), Tang Scholar (JX), and Startup Research Funding of Fudan University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, J., Brosseau, JP. & Shi, H. Targeted degradation of immune checkpoint proteins: emerging strategies for cancer immunotherapy. Oncogene 39, 7106–7113 (2020). https://doi.org/10.1038/s41388-020-01491-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01491-w
This article is cited by
-
Clinically relevant stratification of lung squamous carcinoma patients based on ubiquitinated proteasome genes for 3P medical approach
EPMA Journal (2024)
-
Construction and validation of a novel lysosomal signature for hepatocellular carcinoma prognosis, diagnosis, and therapeutic decision-making
Scientific Reports (2023)
-
Circular RNA MTCL1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1QBP ubiquitin degradation and mediating beta-catenin activation
Molecular Cancer (2022)
-
Targeting the immune checkpoint B7-H3 for next-generation cancer immunotherapy
Cancer Immunology, Immunotherapy (2022)
-
Targeting lysosomes in human disease: from basic research to clinical applications
Signal Transduction and Targeted Therapy (2021)