Abstract
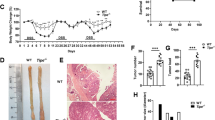
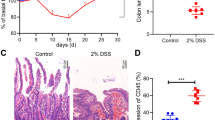
The myeloid translocation gene family member MTG16 is a transcriptional corepressor that relies on the DNA-binding ability of other proteins to determine specificity. One such protein is the ZBTB family member Kaiso, and the MTG16:Kaiso interaction is necessary for repression of Kaiso target genes, such as matrix metalloproteinase-7. Using the azoxymethane and dextran sodium sulfate (AOM/DSS) murine model of colitis-associated carcinoma, we previously determined that MTG16 loss accelerates tumorigenesis and inflammation. However, it was unknown whether this effect was modified by Kaiso-dependent transcriptional repression. To test for a genetic interaction between MTG16 and Kaiso in inflammatory carcinogenesis, we subjected single and double knockout (DKO) mice to the AOM/DSS protocol. Mtg16−/− mice demonstrated increased colitis and tumor burden; in contrast, disease severity in Kaiso−/− mice was equivalent to wild-type controls. Surprisingly, Kaiso deficiency in the context of MTG16 loss reversed injury and pro-tumorigenic responses in the intestinal epithelium following AOM/DSS treatment, and tumor numbers were returned to near to wild-type levels. Transcriptomic analysis of non-tumor colon tissue demonstrated that changes induced by MTG16 loss were widely mitigated by concurrent Kaiso loss, and DKO mice demonstrated downregulation of metabolism and cytokine-associated gene sets with concurrent activation of DNA damage checkpoint pathways as compared with Mtg16−/−. Further, Kaiso knockdown in intestinal enteroids reduced stem- and WNT-associated phenotypes, thus abrogating the induction of these pathways observed in Mtg16−/− samples. Together, these data suggest that Kaiso modifies MTG16-driven inflammation and tumorigenesis and suggests that Kaiso deregulation contributes to MTG16-dependent colitis and CAC phenotypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
08 May 2019
In the original version of this article the authors noted that the GEO accession number for the relevant dataset was listed incorrectly as GSE12454.
References
Daniel JM, Reynolds AB. The cateninp120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–23.
Spring CM, Kelly KF, O’Kelly I, Graham M, Crawford HC, Daniel JM. The catenin p120ctn inhibits Kaiso-mediated transcriptional repression of the beta-catenin/TCF target gene matrilysin. Exp Cell Res. 2005;305:253–65.
Lopes EC, Valls E, Figueroa ME, Mazur A, Meng FG, Chiosis G, et al. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res. 2008;68:7258–63.
Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–81.
Buck-Koehntop BA, Martinez-Yamout MA, Dyson HJ, Wright PE. Kaiso uses all three zinc fingers and adjacent sequence motifs for high affinity binding to sequence-specific and methyl-CpG DNA targets. FEBS Lett. 2012;586:734–9.
Donaldson IJ, Amin S, Hensman JJ, Kutejova E, Rattray M, Lawrence N, et al. Genome-wide occupancy links Hoxa2 to Wnt-beta-catenin signaling in mouse embryonic development. Nucleic Acids Res. 2012;40:3990–4001.
Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–54.
Pozner A, Terooatea TW, Buck-Koehntop BA. Cell-specific Kaiso (ZBTB33) regulation of cell cycle through cyclin D1 and cyclin E1. J Biol Chem. 2016;291:24538–50.
Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ, et al. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136:729–38.
Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10.
Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–84.
Barrett CW, Smith JJ, Lu LC, Markham N, Stengel KR, Short SP, et al. Kaiso directs the transcriptional corepressor MTG16 to the Kaiso binding site in target promoters. PLoS ONE. 2012;7:e51205.
Kochetkova M, McKenzie OL, Bais AJ, Martin JM, Secker GA, Seshadri R, et al. CBFA2T3 (MTG16) is a putative breast tumor suppressor gene from the breast cancer loss of heterozygosity region at 16q24.3. Cancer Res. 2002;62:4599–604.
Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74.
Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13.
Barrett CW, Fingleton B, Williams A, Ning W, Fischer MA, Washington MK, et al. MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res. 2011;71:1302–12.
Pierre CC, Longo J, Mavor M, Milosavljevic SB, Chaudhary R, Gilbreath E, et al. Kaiso overexpression promotes intestinal inflammation and potentiates intestinal tumorigenesis in Apc(Min/+) mice. Biochim Biophys Acta. 2015;1852:1846–55.
Chaudhary R, Pierre CC, Nanan K, Wojtal D, Morone S, Pinelli C, et al. The POZ-ZF transcription factor Kaiso (ZBTB33) induces inflammation and progenitor cell differentiation in the murine intestine. PLoS ONE. 2013;8:e74160.
Prokhortchouk A, Sansom O, Selfridge J, Caballero IM, Salozhin S, Aithozhina D, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208.
The Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7.
McDonough EM, Barrett CW, Parang B, Mittal MK, Smith JJ, Bradley AM, et al. MTG16 is a tumor suppressor in colitis-associated carcinoma. JCI Insight. 2017;2.
Becker C, Fantini MC, Wirtz S, Nikolaev A, Kiesslich R, Lehr HA, et al. In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut. 2005;54:950–4.
Williams CS, Bradley AM, Chaturvedi R, Singh K, Piazuelo MB, Chen X. MTG16 contributes to colonic epithelial integrity in experimental colitis. Gut. 2012;62:1446–55.
Poindexter SV, Reddy VK, Mittal MK, Williams AM, Washington MK, Harris E, et al. Transcriptional corepressor MTG16 regulates small intestinal crypt proliferation and crypt regeneration after radiation-induced injury. Am J Physiol Gastrointest Liver Physiol. 2015;308:G562–71.
Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, et al. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. 2015;163:1444–56.
Bergstrom KS, Guttman JA, Rumi M, Ma C, Bouzari S, Khan MA, et al. Modulation of intestinal goblet cell function during infection by an attaching and effacing bacterial pathogen. Infect Immun. 2008;76:796–811.
McCormick DA, Horton LW, Mee AS. Mucin depletion in inflammatory bowel disease. J Clin Pathol. 1990;43:143–6.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40.
Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–34.
Syljuasen RG, Krolewski B, Little JB. Loss of normal G1 checkpoint control is an early step in carcinogenesis, independent of p53 status. Cancer Res. 1999;59:1008–14.
Rudolph KL, Hartmann D, Opitz OG. Telomere dysfunction and DNA damage checkpoints in diseases and cancer of the gastrointestinal tract. Gastroenterology. 2009;137:754–62.
Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS, et al. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Mol Cell. 2012;46:335–50.
Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–45.
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5.
Reddy VK, Short SP, Barrett CW, Mittal MK, Keating CE, Thompson JJ, et al. BVES regulates intestinal stem cell programs and intestinal crypt viability after radiation. Stem Cells. 2016;34:1626–36.
Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–82.
Kim SW, Park JI, Spring CM, Sater AK, Ji H, Otchere AA, et al. Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat Cell Biol. 2004;6:1212–20.
Li Q, Ishikawa TO, Oshima M, Taketo MM. The threshold level of adenomatous polyposis coli protein for mouse intestinal tumorigenesis. Cancer Res. 2005;65:8622–7.
Buchert M, Athineos D, Abud HE, Burke ZD, Faux MC, Samuel MS, et al. Genetic dissection of differential signaling threshold requirements for the Wnt/beta-catenin pathway in vivo. PLoS Genet. 2010;6:e1000816.
Christie M, Jorissen RN, Mouradov D, Sakthianandeswaren A, Li S, Day F, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/beta-catenin signalling thresholds for tumourigenesis. Oncogene. 2013;32:4675–82.
Koch S, Nava P, Addis C, Kim W, Denning TL, Li L, et al. The Wnt antagonist Dkk1 regulates intestinal epithelial homeostasis and wound repair. Gastroenterology. 2011;141:259–68, 68 e1–8.
Martinez JA, Williams CS, Amann JM, Ellis TC, Moreno-Miralles I, Washington MK, et al. Deletion of Mtgr1 sensitizes the colonic epithelium to dextran sodium sulfate-induced colitis. Gastroenterology. 2006;131:579–88.
Blattler A, Yao L, Wang Y, Ye Z, Jin VX, Farnham PJ. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin. 2013;6:13.
Becker C, Fantini MC, Neurath MF. High resolution colonoscopy in live mice. Nat Protoc. 2006;1:2900–4.
Brown JJ, Short SP, Stencel-Baerenwald J, Urbanek K, Pruijssers AJ, McAllister N, et al. Reovirus-Induced Apoptosis in the Intestine Limits Establishment of Enteric Infection. J Virol. 2018;92.
Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M, et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 2013;3:1128–39.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.
Acknowledgements
The authors thank members of the Williams and Reynolds laboratories for thoughtful discussions about this research project. We would also like to thank the Vanderbilt Translational Pathology Shared Resource for aid with histology and the Vanderbilt Technologies for Advanced Genomics for next gen sequencing.
Funding
National Institutes of Health (R01DK099204 to CSW; R01AT004821 to KTW; R01CA178030 to SWH; 1F31CA167920 to CWB; F32DK108492 to SPS); the Federal Research Center “Fundamentals of Biotechnology” budget (No. 01201371085 to EP); Office of Medical Research, Department of Veterans Affairs (1I01BX001426 to CSW; 1I01BX001453 to KTW; 1IK2BX002126 to LAC). Additional support was provided by the NIH grant P30DK058404 (Vanderbilt Digestive Disease Research Center).
Author information
Authors and Affiliations
Contributions
SPS, CWB, KRS, KTW, EP, SWH, ABR and CSW: study conception and/or design; SPS, CWB, KRS, FLR, YAC, LAC, MKL and EMM: data acquisition; SPS, CWB, KRS, YAC, MKW, XC and CSW: data analysis; SPS, CWB, LAC, MKW, KTW, EP and SWH: financial support. All authors contributed to paper generation and approval.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Short, S.P., Barrett, C.W., Stengel, K.R. et al. Kaiso is required for MTG16-dependent effects on colitis-associated carcinoma. Oncogene 38, 5091–5106 (2019). https://doi.org/10.1038/s41388-019-0777-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0777-7