Abstract
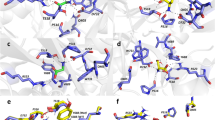
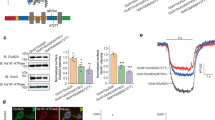
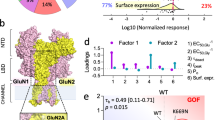
NMDA receptors have essential roles in the physiology of central excitatory synapses and their dysfunction causes severe neuropsychiatric symptoms. Recently, a series of genetic variants have been identified in patients, however, functional information about these variants is sparse and their role in pathogenesis insufficiently known. Here we investigate the mechanism by which two GluN2A variants may be pathogenic. We use molecular dynamics simulation and single-molecule electrophysiology to examine the contribution of GluN2A subunit-residues, P552 and F652, and their pathogenic substitutions, P552R and F652V, affect receptor functions. We found that P552 and F652 interact during the receptors’ normal activity cycle; the interaction stabilizes receptors in open conformations and is required for a normal electrical response. Engineering shorter side-chains at these positions (P552A and/or F652V) caused a loss of interaction energy and produced receptors with severe gating, conductance, and permeability deficits. In contrast, the P552R side chain resulted in stronger interaction and produced a distinct, yet still drastically abnormal electrical response. These results identify the dynamic contact between P552 and F652 as a critical step in the NMDA receptor activation, and show that both increased and reduced communication through this interaction cause dysfunction. Results show that subtle differences in NMDA receptor primary structure can generate complex phenotypic alterations whose binary classification is too simplistic to serve as a therapeutic guide.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fernandez-Marmiesse A, Kusumoto H, Rekarte S, Roca I, Zhang J, Myers SJ, et al. A novel missense mutation in GRIN2A causes a nonepileptic neurodevelopmental disorder. Mov Disord. 2018;33:992–9.
Lemke JR, Lal D, Reinthaler EM, Steiner I, Nothnagel M, Alber M, et al. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat Genet. 2013;45:1067–72.
Carvill GL, Regan BM, Yendle SC, O’Roak BJ, Lozovaya N, Bruneau N, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013;45:1073–6.
de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9.
Lesca G, Rudolf G, Bruneau N, Lozovaya N, Labalme A, Boutry-Kryza N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061–6.
Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–6.
Tarabeux J, Kebir O, Gauthier J, Hamdan FF, Xiong L, Piton A, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry. 2011;1:e55.
Garcia-Recio A, Santos-Gomez A, Soto D, Julia-Palacios N, Garcia-Cazorla A, Altafaj X, et al. GRIN database: a unified and manually curated repertoire of GRIN variants. Hum Mutat. 2021;42:8–18.
Lemke JR, Geider K, Helbig KL, Heyne HO, Schutz H, Hentschel J, et al. Delineating the GRIN1 phenotypic spectrum: a distinct genetic NMDA receptor encephalopathy. Neurology. 2016;86:2171–8.
Burnashev N, Szepetowski P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharm. 2015;20:73–82.
Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF. Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol Pharm. 2015;88:203–17.
Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, et al. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13:325–38.
Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S. Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell. 1994;76:427–37.
Sprengel R, Single FN. Mice with genetically modified NMDA and AMPA receptors. Ann N. Y Acad Sci. 1999;868:494–501.
Fayyazuddin A, Villarroel A, Le Goff A, Lerma J, Neyton J. Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron. 2000;25:683–94.
Amico-Ruvio SA, Paganelli MA, Abbott JA, Myers JM, Kasperek EM, Iacobucci GI et al. Contributions by N-terminal Domains to NMDA Receptor Currents. bioRxiv 2020: 2020.2008.2021.261388. https://doi.org/10.1101/2020.08.21.261388.
Maki BA, Aman TK, Amico-Ruvio SA, Kussius CL, Popescu GK. C-terminal domains of N-methyl-D-aspartic acid receptor modulate unitary channel conductance and gating. J Biol Chem. 2012;287:36071–80.
Sprengel R, Suchanek B, Amico C, Brusa R, Burnashev N, Rozov A, et al. Importance of the intracellular domain of NR2 subunits for NMDA receptor function in vivo. Cell. 1998;92:279–89.
Pierson TM, Yuan H, Marsh ED, Fuentes-Fajardo K, Adams DR, Markello T, et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol. 2014;1:190–8.
Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, et al. Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet. 2017;13:e1006536.
Amin JB, Leng X, Gochman A, Zhou HX, Wollmuth LP. A conserved glycine harboring disease-associated mutations permits NMDA receptor slow deactivation and high Ca(2+) permeability. Nat Commun. 2018;9:3748.
Yuan H, Hansen KB, Zhang J, Pierson TM, Markello TC, Fajardo KV, et al. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun. 2014;5:3251.
Fedele L, Newcombe J, Topf M, Gibb A, Harvey RJ, Smart TG. Disease-associated missense mutations in GluN2B subunit alter NMDA receptor ligand binding and ion channel properties. Nat Commun. 2018;9:957.
Li D, Yuan H, Ortiz-Gonzalez XR, Marsh ED, Tian L, McCormick EM, et al. GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am J Hum Genet. 2016;99:802–16.
Chen W, Shieh C, Swanger SA, Tankovic A, Au M, McGuire M, et al. GRIN1 mutation associated with intellectual disability alters NMDA receptor trafficking and function. J Hum Genet. 2017;62:589–97.
Serraz B, Grand T, Paoletti P. Altered zinc sensitivity of NMDA receptors harboring clinically-relevant mutations. Neuropharmacology. 2016;109:196–204.
Marwick KFM, Parker P, Skehel P, Hardingham G, Wyllie DJA. Functional assessment of the NMDA receptor variant GluN2A (R586K). Wellcome Open Res. 2017;2:20.
Marwick KFM, Hansen KB, Skehel PA, Hardingham GE, Wyllie DJA. Functional assessment of triheteromeric NMDA receptors containing a human variant associated with epilepsy. J Physiol. 2019;597:1691–704.
Marwick K, Skehel P, Hardingham G, Wyllie D. Effect of a GRIN2A de novo mutation associated with epilepsy and intellectual disability on NMDA receptor currents and Mg(2+) block in cultured primary cortical neurons. Lancet. 2015;385:S65.
Iacobucci GJ, Popescu GK. NMDA receptors: linking physiological output to biophysical operation. Nat Rev Neurosci. 2017;18:236–49.
Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, et al. Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol. 2018;150:1081–105.
Elmasri M, Hunter DW, Winchester G, Bates EE, Aziz W, Van Der Does DM, et al. Common synaptic phenotypes arising from diverse mutations in the human NMDA receptor subunit GluN2A. Commun Biol. 2022;5:174.
Elmasri M, Lotti JS, Aziz W, Steele OG, Karachaliou E, Sakimura K et al. Synaptic Dysfunction by Mutations in GRIN2B: Influence of Triheteromeric NMDA Receptors on Gain-of-Function and Loss-of-Function Mutant Classification. Brain Sci. 2022;12:789–813.
Iacobucci GJ, Wen H, Helou M, Liu B, Zheng W, Popescu GK. Cross-subunit interactions that stabilize open states mediate gating in NMDA receptors. Proc Natl Acad Sci USA. 2021;118:e2007511118.
Longo PA, Kavran JM, Kim MS, Leahy DJ. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 2013;529:227–40.
Barry PH, Lynch JW. Liquid junction potentials and small cell effects in patch-clamp analysis. J Membr Biol. 1991;121:101–17.
Lewis CA. Ion-concentration dependence of the reversal potential and the single channel conductance of ion channels at the frog neuromuscular junction. J Physiol. 1979;286:417–45.
Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J. 2004;86:1488–501.
Qin F, Auerbach A, Sachs F. Maximum likelihood estimation of aggregated Markov processes. Proc Biol Sci. 1997;264:375–83.
Kussius CL, Kaur N, Popescu GK. Pregnanolone sulfate promotes desensitization of activated NMDA receptors. J Neurosci. 2009;29:6819–27.
Popescu G, Robert A, Howe JR, Auerbach A. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–3.
Zheng W, Wen H, Iacobucci GJ, Popescu GK. Probing the Structural Dynamics of the NMDA Receptor Activation by Coarse-Grained Modeling. Biophys J. 2017;112:2589–601.
Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–7.
Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, et al. Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature. 2016;534:63–8.
Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–60. 376
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802.
Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27-38
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–985.
Gibb AJ, Ogden KK, McDaniel MJ, Vance KM, Kell SA, Butch C, et al. A structurally derived model of subunit-dependent NMDA receptor function. J Physiol. 2018;596:4057–89.
Horovitz A. Double-mutant cycles: a powerful tool for analyzing protein structure and function. Fold Des. 1996;1:R121–R126.
Maki BA, Cummings KA, Paganelli MA, Murthy SE, Popescu GK. One-channel cell-attached patch-clamp recording. J Vis Exp. 2014;88:e51629.
Amico-Ruvio SA, Popescu GK. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys J. 2010;98:1160–9.
Cummings KA, Iacobucci GJ, Popescu GK. Extracting Rate Constants for NMDA Receptor Gating from One-Channel Current Recordings. In: Popescu GK, editor. Ionotropic Glutamate Receptor Technologies, 106. New York:Springer; 2016. p. 273–99.
Chowdhury S, Haehnel BM, Chanda B. A self-consistent approach for determining pairwise interactions that underlie channel activation. J Gen Physiol. 2014;144:441–55.
Popescu G, Auerbach A. Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci. 2003;6:476–83.
Maki BA, Popescu GK. Extracellular Ca(2+) ions reduce NMDA receptor conductance and gating. J Gen Physiol. 2014;144:379–92.
Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–96.
Amin JB, Gochman A, He M, Certain N, Wollmuth LP. NMDA Receptors Require Multiple Pre-opening Gating Steps for Efficient Synaptic Activity. Neuron. 2021;109:488–501.e484.
Lau AY, Salazar H, Blachowicz L, Ghisi V, Plested AJ, Roux B. A conformational intermediate in glutamate receptor activation. Neuron. 2013;79:492–503.
Baranovic J, Chebli M, Salazar H, Carbone AL, Faelber K, Lau AY, et al. Dynamics of the Ligand Binding Domain Layer during AMPA Receptor Activation. Biophys J. 2016;110:896–911.
Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–34.
Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat Neurosci. 2014;17:914–22.
Balannik V, Menniti FS, Paternain AV, Lerma J, Stern-Bach Y. Molecular mechanism of AMPA receptor noncompetitive antagonism. Neuron. 2005;48:279–88.
Perszyk R, Katzman BM, Kusumoto H, Kell SA, Epplin MP, Tahirovic YA et al. An NMDAR positive and negative allosteric modulator series share a binding site and are interconverted by methyl groups. Elife. 2018;7:e34711.
Perszyk RE, Swanger SA, Shelley C, Khatri A, Fernandez-Cuervo G, Epplin MP, et al. Biased modulators of NMDA receptors control channel opening and ion selectivity. Nat Chem Biol. 2020;16:188–96.
Watanabe J, Beck C, Kuner T, Premkumar LS, Wollmuth LP. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci. 2002;22:10209–16.
Talukder I, Borker P, Wollmuth LP. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci. 2010;30:11792–804.
Schmid SM, Korber C, Herrmann S, Werner M, Hollmann M. A domain linking the AMPA receptor agonist binding site to the ion pore controls gating and causes lurcher properties when mutated. J Neurosci. 2007;27:12230–41.
Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous Movements Prior to Pore Opening in NMDA Receptors. J Neurosci. 2013;33:12052–66.
Tian N, Boring M, Kobau R, Zack MM, Croft JB. Active Epilepsy and Seizure Control in Adults - United States, 2013 and 2015. MMWR Morb Mortal Wkly Rep. 2018;67:437–42.
Kalilani L, Sun X, Pelgrims B, Noack-Rink M, Villanueva V. The epidemiology of drug-resistant epilepsy: a systematic review and meta-analysis. Epilepsia. 2018;59:2179–93.
Strehlow V, Heyne HO, Vlaskamp DRM, Marwick KFM, Rudolf G, de Bellescize J, et al. GRIN2A-related disorders: genotype and functional consequence predict phenotype. Brain. 2019;142:80–92.
McDaniel MJ, Ogden KK, Kell SA, Burger PB, Liotta DC, Traynelis SF. NMDA receptor channel gating control by the pre-M1 helix. J Gen Physiol. 2020;152:e201912362.
Kaniakova M, Kleteckova L, Lichnerova K, Holubova K, Skrenkova K, Korinek M, et al. 7-Methoxyderivative of tacrine is a ‘foot-in-the-door’ open-channel blocker of GluN1/GluN2 and GluN1/GluN3 NMDA receptors with neuroprotective activity in vivo. Neuropharmacology. 2018;140:217–32.
Popescu G. Principles of N-methyl-D-aspartate receptor allosteric modulation. Mol Pharm. 2005;68:1148–55.
Glasgow NG, Povysheva NV, Azofeifa AM, Johnson JW. Memantine and Ketamine Differentially Alter NMDA Receptor Desensitization. J Neurosci. 2017;37:9686–704.
Popescu G. Mechanism-based targeting of NMDA receptor functions. Cell Mol Life Sci. 2005;62:2100–11.
Acknowledgements
We thank Jamie Abbott, PhD for helpful critiques and technical support in cell culture and molecular biology. This study was funded by NIH R01NS108750 and R01NS097016 to GKP.
Author information
Authors and Affiliations
Contributions
GJI and GKP conceived the study. HW and WZ performed and analyzed MD simulations. GJI, BL, and BS performed and analyzed electrophysiology experiments. GJI, BL, and GKP prepared figures and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iacobucci, G.J., Liu, B., Wen, H. et al. Complex functional phenotypes of NMDA receptor disease variants. Mol Psychiatry 27, 5113–5123 (2022). https://doi.org/10.1038/s41380-022-01774-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01774-6
This article is cited by
-
GRIN2A (NR2A): a gene contributing to glutamatergic involvement in schizophrenia
Molecular Psychiatry (2023)