Abstract
DNA methylation profiles of aggressive behavior may capture lifetime cumulative effects of genetic, stochastic, and environmental influences associated with aggression. Here, we report the first large meta-analysis of epigenome-wide association studies (EWAS) of aggressive behavior (N = 15,324 participants). In peripheral blood samples of 14,434 participants from 18 cohorts with mean ages ranging from 7 to 68 years, 13 methylation sites were significantly associated with aggression (alpha = 1.2 × 10−7; Bonferroni correction). In cord blood samples of 2425 children from five cohorts with aggression assessed at mean ages ranging from 4 to 7 years, 83% of these sites showed the same direction of association with childhood aggression (r = 0.74, p = 0.006) but no epigenome-wide significant sites were found. Top-sites (48 at a false discovery rate of 5% in the peripheral blood meta-analysis or in a combined meta-analysis of peripheral blood and cord blood) have been associated with chemical exposures, smoking, cognition, metabolic traits, and genetic variation (mQTLs). Three genes whose expression levels were associated with top-sites were previously linked to schizophrenia and general risk tolerance. At six CpGs, DNA methylation variation in blood mirrors variation in the brain. On average 44% (range = 3–82%) of the aggression–methylation association was explained by current and former smoking and BMI. These findings point at loci that are sensitive to chemical exposures with potential implications for neuronal functions. We hope these results to be a starting point for studies leading to applications as peripheral biomarkers and to reveal causal relationships with aggression and related traits.
Similar content being viewed by others
Introduction
Aggression encompasses a range of behaviors, such as bullying, verbal abuse, fighting, and destroying objects. Early life social conditions, including low parental income, separation from a parent, family dysfunction, and maternal smoking during pregnancy are risk factors for childhood aggression [1,2,3]. High levels of aggression are a characteristic of several psychiatric disorders and may also be caused by traumatic brain injury [3], neurodegenerative diseases [4] and alcohol and substance abuse [5, 6].
DNA methylation mediates effects of genetic variants in regulatory regions on gene expression [7] and is modifiable by early life social environment, as demonstrated by animal studies [8, 9], and by chemical exposures including (prenatal) exposure to cigarette smoke, as illustrated by numerous human studies [10]. Despite the large tissue-specificity of DNA methylation, effects of genetic variants on nearby DNA methylation (cis mQTLs) correlate strongly between blood and brain cells [11]. DNA methylation signatures of chemical exposures [12] and maternal rearinging [9] show a certain (but less understood) degree of conservation across tissues.
Large-scale epigenome-wide association studies (EWASs) have become feasible through DNA methylation microarrays applied to blood samples from large cohorts, identifying thousands of loci where methylation in cord blood is associated with maternal smoking [13]. Methylation in blood is associated with depressive symptoms [14] and brain morphology [15], with some evidence for blood DNA methylation signatures being a marker for methylation levels [15] or gene expression [14] in the brain. For several traits, DNA methylation scores based on multiple CpGs from EWAS show better predictive value than currently available polygenic scores [16, 17].
Small-scale studies (maximum sample size = 260) have provided some evidence that DNA methylation differences in blood, cord blood, and buccal cells are associated with severe forms of aggressive behavior and related problems in children and adults, including (chronic) physical aggression and early onset conduct problems [18,19,20], but studies on violent aggression in schizophrenia patients (N = 134) [21] and a population-based study of continuous aggression symptoms in adults (N = 2029) [22] did not detect epigenome-wide significant sites.
We performed an EWAS meta-analysis of aggressive behavior and closely related constructs. We chose to meta-analyze multiple measures of aggression across ages and sex to maximize sample size. The contribution of genetic influences to aggression is largely stable, at least throughout childhood [23], whereas epigenetic signatures may be dynamic and may differ across cell types and age. Therefore, we performed separate meta-analyses of peripheral blood collected after birth (N = 14,434) and cord blood (N = 2425), followed by a combined meta-analysis (N = 15,324) including an examination of heterogeneity of effects. Next, we tested the relationship between aggressive behavior and epigenetic clocks, as associations of lifetime stress [24], exposure to violence [25], and psychiatric disorders [26, 27] with accelerated epigenetic ageing have been reported. We performed extensive functional follow-up by integrating our findings with data on gene expression, mQTLs and DNA methylation in brain samples.
Methods
Cohorts
Demographic information for the cohorts is provided in Table 1. Detailed cohort information is provided in eAppendix 1. Informed consent was obtained from all participants. The protocol for each study was approved by the ethical review board of each institution.
Aggressive behavior
Aggressive behavior was assessed by self-report or reported by parents and teachers. Multiple instruments were used (eTable 1): ASEBA Child Behavior Check List (CBCL) [28], Strengths and Difficulties Questionnaire (SDQ) conduct problem scale [29], Multidimensional Peer Nomination Inventory (MNPI) aggression scale [30], ASEBA adult self-report (ASR) aggression scale [31], DSM-IV Conduct Disorder Symptom Scale [32], Multidimensional Personality Questionnaire (MPQ) aggression scale [33], and the Hunter–Wolf aggressive behavior scale [34, 35]. In four cohorts, a single aggression-related item from personality questionnaires was used. Distributions of aggression scores are provided in eFig. 1.
DNA methylation BeadChips
DNA methylation was assessed with Illumina BeadChips: the llumina Infinium HumanMethylation450 BeadChip (450k array; majority of cohorts), or the Illumina MethylationEPIC BeadChip (EPIC array). Most cohorts analyzed DNA methylation β-values, which range from 0 to 1, indicating the proportion of DNA that is methylated at a CpG in a sample. Cohort-specific details about DNA methylation profiling, quality control, and normalization are described in eAppendix 1 and summarized in eTable 2.
Epigenome-wide association analysis
EWAS analyses were performed according to a standard operating procedure (http://www.action-euproject.eu/content/data-protocols). In each cohort, the association between DNA methylation level and aggressive behavior was specified under a linear model with DNA methylation as outcome, and correction for relatedness of individuals where applicable. Two models were tested. Model 1 included aggressive behavior, sex, age at blood sampling (not in cohorts with invariable age), white blood cell percentages (measured or imputed), and technical covariates. Model 2 included the same predictors plus body-mass-index (BMI) and smoking status in adolescents and adults (current smoker, former smoker or never smoked). Cohort-specific details and R-code are provided in eAppendix 1 and eTable 3, respectively. The relationship between aggressive behavior and covariates is provided in eTable 4 based on data from the Netherlands Twin Register (N = 2059).
Quality control and filtering of cohort-level EWAS summary statistics is described in eAppendix 2. The following probes were removed: on sex chromosomes, methylation sites with more than 5% missing data in a cohort, probes overlapping SNPs affecting the CpG or single base extension site with a minor allele frequency (MAF) > 0.01 in the 1000 G EU or GONL population [7], and ambiguous mapping probes reported with an overlap of at least 47 bases per probe [36]. The R package Bacon was used to compute the Bayesian inflation factor and to obtain bias- and inflation-corrected test statistics (eFig. 2) prior to meta-analysis [37]. Further data can be found in the supplementary material for this paper, eFigs. 1–18
Meta-analysis
Fixed-effects meta-analyses were performed in METAL [38]. We used the p-value-based (sample size-weighted) method because the measurement scale of aggressive behavior differs across studies. First, results based on peripheral blood and cord blood data were meta-analyzed separately. Second, a combined meta-analysis was performed of all data. The following cohorts had data available for both cord blood and peripheral blood (from the same children): INMA (which is part of HELIX) and ALSPAC. In the combined meta-analysis, the cord blood data from ALSPAC and INMA were excluded to avoid sample overlap. Statistical significance was assessed considering Bonferroni correction for the number of sites tested (alpha = 1.2 × 10−7). Methylation sites that were associated with aggression at the less conservative false discovery rate (FDR) threshold (5%) were included in follow-up analyses. The I2 statistic from METAL was used to describe heterogeneity.
Follow-up analyses
DNA methylation score analyses and epigenetic clock analyses are described in eAppendix 3 and eAppendix 4. Follow-up analyses (eAppendix 5- eAppendix 10) were performed on meta-analysis top-sites (FDR < 0.05), including a comparison of top-sites with all previously reported associations in the EWAS atlas [39], follow-up analysis of top-sites in two clinical cohorts with blood methylation data (Table 2), a cross-tissue analysis (blood, buccal, brain), and association with gene expression level and mQTLs. Analyses of differentially methylated regions (DMRs) are described in eAppendix 8. Finally, we performed replication analysis of a previously reported DMR associated with aggression [20] (eAppendix 9).
Results
Peripheral blood meta-analysis
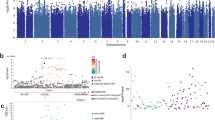
We performed a meta-analysis of 13 studies with peripheral blood DNA methylation data (N = 14,434). The meta-analysis test statistics showed no inflation (eTable 5, eFig. 3). In model 1, methylation at 13 CpGs was associated with aggression (Bonferroni correction; alpha = 1.2 × 10−7), and 35 passed a less conservative threshold (FDR 5%; Fig. 1a). At 28 out of the 35 sites (80%), higher levels of aggression were associated with lower methylation levels. Top-sites showed varying degrees of between-study heterogeneity (mean I2 = 50%; range = 0–86%, eTable 6). Five sites showed significant heterogeneity (alpha = 1.2 × 10−7).
a Manhattan plot showing the fixed effects meta-analysis p values for the association between aggressive behavior and DNA methylation level based on the meta-analysis of peripheral blood. The blue horizontal line denotes the FDR-threshold (5%) and the red line indicates the Bonferroni threshold. b Effects sizes of top-sites from the meta-analysis of aggression in peripheral blood (x-axis) versus effects sizes from the meta-analysis of aggression in cord blood (y-axis). c Venn diagram showing the numbers and overlap of CpGs detected at FDR 5% in the meta-analysis of peripheral blood and the combined meta-analysis and cord blood and peripheral blood. d Effects sizes of top-sites from the meta-analysis of aggression in peripheral blood model 1 (x-axis) versus effects sizes from the meta-analysis of aggression in peripheral blood model 2; adjusted for smoking and BMI (y-axis). e Top enriched traits based on enrichment analysis with all 48 top-sites. The third column shows how many of the 48 CpGs have been previously associated with the trait in the first column. The last column shows the overlap as a percentage of the total number of CpGs previously associated with the trait in column 1 (e.g. 0.3% of all CpGs previously associated with smoking are also associated with aggression in the current meta-analysis). d In b and d, CpGs that have not been previously associated with smoking in the meta-analysis by Joehanes et al. [40] are plotted in red.
Cord blood meta-analysis
The meta-analysis of cord blood (five cohorts; N = 2425) detected no significant CpGs (eTable 7). Examining top-sites from the peripheral blood meta-analysis, 12 of the significant, and 33 of the FDR top-sites were assessed in cord blood; 10 (83%), and 25 (71%), respectively, showed the same direction of association (Fig. 1b). Effect sizes in cord blood correlated significantly with effect sizes in peripheral blood (r = 0.74, p = 0.006 for epigenome-wide significant and r = 0.51, p = 0.003 for FDR top-sites).
Combined meta-analysis
In the combined meta-analysis of peripheral and cord blood data (total sample size = 15,324, eTable 6), methylation at 13 CpGs was associated with aggression after Bonferroni correction, including ten CpGs from the peripheral blood meta-analysis, and 43 passed a less conservative threshold (FDR 5%, Table 3). Among FDR top-sites from both analyses, 13 CpGs were only found in the combined meta-analysis but not in the peripheral blood meta-analysis, while five CpGs from the peripheral blood meta-analysis were no longer significant in the combined meta-analysis (Fig. 1c).
CBCL meta-analysis
We compared our meta-analysis results to a meta-analysis of cohorts that applied the same aggression instrument; i.e. CBCL (four studies; N = 2286; Table 1). No epigenome-wide significant sites were detected (eFig. 4a). Examining top-sites from the overall meta-analysis (Model 1), 38 (79%) showed the same direction of association for CBCL aggression in children, and effect sizes correlated strongly (r = 0.75, p = 6.8 × 10−10, eFig. 4b).
Overlap with CpGs detected in previous EWASs
We performed enrichment analyses against all previously reported associations with diseases and environmental exposures recorded in the EWAS Atlas [39]. The top ten most strongly enriched traits are shown in Fig. 1e. CpGs associated with aggressive behavior showed large overlap with CpGs previously associated with smoking (37 CpGs; corresponding to 77% of aggression-associated CpGs and 0.3% of CpGs that have been previously associated with smoking), and smaller overlap with other smoking traits (e.g. maternal smoking), other chemical exposures (e.g. perinatal exposure to polychlorinated biphenyls (PCBs) and polychlorinated dibenzofurans (PCDFs)). Further overlap includes CpGs associated with alcohol consumption, cognitive function, educational attainment, ageing, and metabolic traits (eTable 8).
Controlling for smoking and BMI
Model 2 was fitted to test whether the association between DNA methylation and aggressive behavior attenuated after adjusting for the most important postnatal lifestyle factors that influence DNA methylation (smoking and BMI). Examining 17,457 CpGs associated with smoking [40], previously reported effect sizes for smoking correlated significantly with effect sizes for aggression from our meta-analysis (r = 0.55, p < 1 × 10−16, eFig. 5a). Examining the 35 CpGs associated with aggression at FDR 5% in peripheral blood, all CpGs showed the same direction of association with aggression after adjusting for smoking and BMI (eTable 6, Fig. 1d). Effect sizes were attenuated to varying degrees (mean reduction = 44%, range = 3–83%). Changes in effect sizes are likely primarily driven by the correction for smoking, since only one top-site has been associated previously with BMI. Some CpGs showed little attenuation, in particular CpGs that have not been previously associated with smoking (e.g.; cg02895948; PLXNA2, cg00891184; KIF1B, cg1215892; intergenic, and cg05432213; ACT1; eFig. 5b). In model 2, between-study heterogeneity at top-sites was greatly reduced (adjusted: mean I2 = 28%, range = 0–77%). No CpGs were epigenome-wide significant or FDR-significant in the adjusted meta-analyses.
DNA methylation scores
We computed weighted sumscores in NTR (peripheral blood, mean age = 36.4, SD = 12, N = 2,059) based on summary statistics from the peripheral blood meta-analysis without NTR (Fig. 2). The best score, based on CpGs with p < 1 × 10−3 in model 2 (745 CpGs), explained 0.29% of the variance in aggression (p = 0.02, not significant after multiple testing correction). This effect was attenuated when age and sex were added to the prediction equation.
The bars indicate how much of the variance in ASEBA adult self-report (ASR) aggression scores were explained by DNA methylation scores in NTR (N = 2059, peripheral blood, 450k array). Scores were created based on weights from the peripheral blood meta-analysis with NTR excluded (N = 12,375). The y-axis shows percentage of variance explained. Different colors denote DNA methylation scores created with different numbers of CpGs that were selected on their p value in the meta-analysis (see legend). From left to right, the first three plots show DNA methylation scores created based on weights obtained from the meta-analysis of EWAS model 1, and plots 4 till 6 show DNA methylation scores created based on weights obtained from the meta-analysis of EWAS model 2. Each DNA methylation score was tested for association with aggression in three model: the simplest model (first plot) included aggression as outcome variable, and DNA methylation score as predictor plus technical covariates and cell counts. The second model additionally included sex and age as predictors. The third model additionally included sex, age, and smoking as predictors. Stars denote nominal p values < 0.05 (not corrected for multiple testing).
Epigenetic clocks
Horvath and Hannum epigenetic age acceleration were not associated with aggression (eTable 9) in a meta-analysis of 12 studies with peripheral blood DNA methylation data (N = 9554), five studies with cord blood DNA methylation (N = 2,225), or in a combined meta-analysis of 15 studies (N = 9740). There was no significant heterogeneity between cohorts (mean I2 = 16%, range = 0–60%).
Follow-up in clinical cohorts
To assess the translation of our observations to aggression-related problem behavior in psychiatric disorders that show comorbidity with aggression, we performed follow-up analyses of top-sites in two clinical cohorts (Table 2): the NeuroIMAGE [41] cohort of ADHD cases and controls (Ntotal = 71) and the FemNAT-CD [42] cohort of female conduct disorder cases and controls (Ntotal = 100). Results did not replicate (eAppendix 6, eTable 10, eTable 11, eFig. 6, eFig. 7).
Cross-tissue analysis
To assess the generalizability of our observations in blood to other tissues, we examined the association with CBCL aggression in buccal DNA methylation data (EPIC array), available for 38 top-sites, in a twin cohort (N = 1237) and a child clinical cohort (N = 172; Table 2, eTable 12) [43]. We also tested associations with maternal smoking and with child nervous system medication (as indexed by the Anatomical Therapeutic Chemical classification system (ATC N-class))
Correlations between DNA methylation levels in blood and buccal cells, based on 450k data from matched samples (N = 22, age = 18 years) [44] were available for 36 of these CpGs. The average correlation was weak (r = 0.25, range = −0.40–0.76). Five CpGs showed a strong correlation between blood and buccal cells (r > 0.5, eTable 13), of which three have been previously associated with (maternal) smoking.
In line with the weak correlation between blood and buccal cell methylation for most top-sites, none of the top-sites was associated with aggression in buccal samples (alpha = 0.001, eTable 14). Regression coefficients based on analyses in buccal cells and blood overall showed no directional consistency (twin cohort: r = 0.03, p = 0.86; concordant direction: 47%, p = 0.87, binomial test, clinical cohort: r = 0.27, p = 0.10; concordant direction: 61%, p = 0.26). Exclusion of ancestry outliers did not change these results (eTable 14). Of the five CpGs with a large blood-buccal correlation, three showed the same direction of association with aggression in buccal cells from twins, four in clinical cases, and one CpG was nominally associated with aggression in buccal samples from twins; cg11554391 (AHRR), rblood-buccal = 0.69, βaggression = −0.0002, p = 0.007.
One CpG was significantly associated with maternal smoking in both cohorts: cg04180046 (MYO1G), NTR: βmaternalsmoking = 0.041, p = 6.0 × 10−6, Curium: βmaternalsmoking = 0.048, p = 7.9 × 10−5 (eTable 14). None of the CpGs was associated with medication use of the child (eTable 14).
We examined the correlation between DNA methylation levels in blood and brain (N = 122) [45] in published DNA methylation data from matched blood samples and four brain regions. Six aggression top-sites (13%) showed significantly correlated DNA methylation levels between blood and one or multiple brain regions: mean r = 0.52; range = 0.45–0.63, alpha = 2.6 × 10−4, eTable 15, eFig. 8), two of which have not been previously associated with smoking or BMI: cg14560430(TRIM71), and cg20673321(ZNF541).
DMRs
DMR analysis showed that 14 DMPs from our combined meta-analysis reside in regions where multiple correlated methylation sites showed evidence for association with aggressive behavior. DMR analysis also detected additional regions that were not significant in DMP analysis (eTable 16- eTable 21). These analyses are described in detail in eAppendix 8.
Replication analysis
A previous EWAS based on Illumina array data detected a significant DMR in DRD4 in buccal cells associated with engagement in physical fights [20]. This locus did not replicate in our meta-analyses or in the two cohorts with buccal methylation data (eTable 22, eAppendix 9).
Gene expression
Based on peripheral blood RNA-seq and DNA methylation data (N = 2101) [7], 17 significant DNA methylation-gene expression associations were identified among 15 CpGs and ten transcripts (Table 3, eTable 23). For most transcripts, a higher methylation level at a CpG site in cis correlated with lower expression (82.4%): cg03935116 and FAM60A, cg00310412 and SEMA7A, cg03707168 and PPP1R15A, cg03636183 and F2RL3, two intergenic CpGs on chromosome 6, where methylation level correlated negatively with expression levels of FLOT1, TUBB, and LINC00243, and six CpGs annotated to AHRR were negatively associated with EXOC3 expression level. Positive correlations were observed between methylation levels at 2 CpGs on chromosome 7 and levels of RP4-647J21.1 (novel transcript, overlapping MYO1G) and between cg02895948 and PLXNA2.
mQTLs
To gain insight into genetic causes of variation underlying top-sites, we obtained whole-blood mQTL data (N = 3841) [7]. In total, 75 mQTL associations were identified among 34 aggression top-sites (70.8%) and 66 SNPs at the experiment-wide threshold applied by the mQTL study FDR < 0.05): 80% were cis mQTLs and 20% were trans mQTLs (eTable 24).
Discussion
We identified 13 epigenome-wide significant sites (Bonferroni corrected) in the meta-analysis of blood and 13 in the combined meta-analysis of blood and cord blood (16 unique sites). We prioritized 48 top-sites (FDR 5%) for follow-up analyses. Methylation level at three top-sites was associated with expression levels of genes that have been previously linked to psychiatric or behavioral traits in GWASs: FLOT1 (schizophrenia [46]), TUBB (schizophrenia) [46], and PLXNA2 (general risk tolerance) [47]. Several other loci have functions in the brain and six CpGs showed correlated methylation levels between blood and brain.
The majority of top-sites (77%) were associated with smoking, 46% were associated with maternal smoking, 25% were associated with alcohol consumption, and 15% were associated with perinatal PCB and PCDF exposure. This overlap of aggression top-sites with smoking and other chemical exposures is noteworthy. Methylation levels of top-sites in the Aryl-Hydrocarbon Receptor Repressor gene AHRR and several other genes are known to be strongly associated with exposure to cigarette smoke [13, 40] and persistent organic pollutants [48]. The best characterized exogenous ligands of the widely expressed Aryl-Hydrocarbon Receptor are environmental contaminants such as benzo[a]pyrene (B[a]P), and TCDD (dioxin), whose neurotoxic and neuroendocrine effects, including disruption of neuronal proliferation, differentiation, and survival, have been well characterized [49]. Human prenatal exposure to B[a]P is associated with delayed mental development, lower IQ, anxiety and attention problems [50]. Research on B[a]P neurotoxicity in adults is scarce but a study on coke oven workers found that occupational B[a]P exposure correlates with reduced monoamine, amino acid and choline neurotransmitter levels and with impaired learning and memory [51].
On average 44% (range = 3–82%) of the aggression–methylation association was explained by current and former smoking and BMI. Our findings do not merely reflect effects of own smoking: 71% of the top-sites showed the same direction for the prospective association of cord blood methylation at birth and aggression in childhood, and 46% have been associated with maternal prenatal smoking. There is a weak observational association between maternal smoking and child aggression [52]. A limitation of our study is that the EWAS analyses did not adjust for prenatal and postnatal second-hand smoking, and did not adjust for smoking intensity and duration or other substance use. Future studies can examine if the link between prenatal maternal smoking and aggression is mediated by DNA methylation.
We found that DNA methylation scores for aggression explained less variation compared to DNA methylation scores for traits such as BMI, smoking, and educational attainment. For these traits, EWASs tended to identify more epigenome-wide significant hits [16, 17]. The variance in aggression explained by DNA methylation scores was in the same order of magnitude as the variance in height explained by DNA methylation scores (based on EWASs of height in smaller samples), i.e. <1% [16]. More research is needed in particular to delineate a causal link between these methylation sites and aggressive behaviour, since our results may also reflect (residual) confounding by (exposure to second-hand) smoking. One approach to address this could be Mendelian Randomization, in which genetic information (SNPs) is used for causal inference of the effect of an exposure (e.g. DNA methylation) on an outcome (e.g., aggression). This approach previously supported a causal effect of maternal smoking-associated methylation sites in blood on various traits and diseases for which well-powered GWASs have been performed, including schizophrenia [53, 54]. For aggressive behavior, the currently available [55] largest GWASs of aggressive behavior included ~16,000 [56] and ~75,000 participants [57], respectively. The GWAS by Ip et al. detected three significant genes in gene-based analysis, but both GWASs did not detect genome-wide significant SNPs and are likely still underpowered. In the future, larger GWASs of aggressive behavior and larger mQTL analyses will allow for powerful Mendelian Randomization for aggression-associated methylation sites.
Strengths and limitations
This is the largest EWAS of aggressive behavior to date. The large sample size was achieved by applying a broad phenotype definition, including participants from multiple countries and all ages in a meta-analysis, and analyzing DNA methylation data from blood. A limitation of this approach is that it reduces power to detect age-, sex-, and symptom-specific effects, and that genetic and environmental backgrounds of different populations, as well as non-identical processing methods of methylation data play a role. A limitation of population-based cohorts and even clinical populations is that individuals with extreme levels of aggressive behavior who cause most societal problems are likely underrepresented. Moreover, some studies used measures that tapped features that overlap with but are not necessarily indicative of aggression (e.g., personality traits, anger, oppositional defiant disorder). Future EWASs that specifically focus on more homogeneous aggression measures are therefore warranted. Our meta-analysis approach may identify a common epigenomic signature of aggression-related problems.
Follow-up analysis in independent datasets indicated that these findings do not generalize strongly to buccal cells, and results did not replicate in two clinical cohorts. These were small, used different aggression measures, and one used a different technology (sequencing) in females only.
Conclusions
We identified associations between aggressive behavior and DNA methylation in blood at CpGs whose methylation level is also associated with exposure to smoking, alcohol consumption, other chemical exposures, and genetic variation. Methylation levels at three top-sites were associated with expression levels of genes that have been previously linked to psychiatric or behavioral traits in GWAS. Our study illustrates both the merit of EWASs based on peripheral tissues to identify environmentally-driven molecular variation associated with behavioral traits and their challenges to tease-out confounders and mediators of the association, and causality. To have full insight into, and to control for confounders in behavioral EWAS meta-analyses (which, in addition to smoking-exposure across the life course likely include other substance-use and socioeconomic conditions throughout life and other, perhaps less obvious ones) is challenging. Future studies, including those that integrate EWAS results for multiple traits and exposures, DNA methylation in multiple tissues, and GWASs of multiple traits are warranted to unravel the utility of our results as peripheral biomarkers for pathological mechanisms in other tissues (such as neurotoxicity) and to unravel possible causal relationships with aggression and related traits. We consider this study to be the starting point for such follow-up studies.
Code availability
The EWAS R-code is provided in eTable 3.
References
Tremblay RE. Physical aggression during early childhood: trajectories and predictors. Pediatrics. 2004. https://doi.org/10.1542/peds.114.1.e43.
Tremblay RE, Vitaro F, Côté SM. Developmental origins of chronic physical aggression: a bio-psycho-social model for the next generation of preventive interventions. Annu Rev Psychol. 2017. https://doi.org/10.1146/annurev-psych-010416-044030.
Tateno A, Jorge RE, Robinson RG. Clinical correlates of aggressive behavior after traumatic brain injury. J Neuropsychiatry Clin Neurosci. 2014. https://doi.org/10.1176/jnp.15.2.155.
Volicer L, Hurley AC. Management of behavioral symptoms in progressive degenerative dementias. J Gerontol Ser A Biol Sci Med Sci. 2003. https://doi.org/10.1093/gerona/58.9.m837.
Moore TM, Stuart GL, Meehan JC, Rhatigan DL, Hellmuth JC, Keen SM. Drug abuse and aggression between intimate partners: a meta-analytic review. Clin Psychol Rev. 2008;28:247–74.
Boles SM, Miotto K. Substance abuse and violence: a review of the literature. Aggress Violent Behav. 2003;8:155–74.
Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–8.
Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54.
Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. 2012. https://doi.org/10.1523/jneurosci.1470-12.2012.
Martin EM, Fry RC. Environmental influences on the epigenome: exposure- associated DNA methylation in human populations. Annu Rev Public Health. 2018. https://doi.org/10.1146/annurev-publhealth-040617-014629.
Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-04558-1.
Tsai PC, Glastonbury CA, Eliot MN, Bollepalli S, Yet I, Castillo-Fernandez JE, et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin Epigenetics. 2018. https://doi.org/10.1186/s13148-018-0558-0.
Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am J Hum Genet. 2016;98:680–96.
Jovanova OS, Nedeljkovic I, Spieler D, Walker RM, Liu C, Luciano M, et al. DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: meta-analysis of multiethnic epigenome-wide studies. JAMA Psychiatry. 2018. https://doi.org/10.1001/jamapsychiatry.2018.1725.
Jia T, Chu C, Liu Y, van Dongen J, Papastergios E, Armstrong NJ, et al. Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes: findings from the ENIGMA Epigenetics Working Group. Mol Psychiatry. 2019:1–12. https://doi.org/10.1038/s41380-019-0605-z.
Shah S, Bonder MJ, Marioni RE, Zhu Z, McRae AF, Zhernakova A, et al. Improving phenotypic prediction by combining genetic and epigenetic associations. Am J Hum Genet. 2015. https://doi.org/10.1016/j.ajhg.2015.05.014.
McCartney DL, Hillary RF, Stevenson AJ, Ritchie SJ, Walker RM, Zhang Q, et al. Epigenetic prediction of complex traits and death. Genome Biol. 2018. https://doi.org/10.1186/s13059-018-1514-1.
Guillemin C, Provençal N, Suderman M, Côté SM, Vitaro F, Hallett M, et al. DNA methylation signature of childhood chronic physical aggression in T cells of both men and women. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0086822.
Cecil CAM, Walton E, Jaffee SR, O’Connor T, Maughan B, Relton CL, et al. Neonatal DNA methylation and early-onset conduct problems: a genome-wide, prospective study. Dev Psychopathol. 2018. https://doi.org/10.1017/S095457941700092X.
Cecil CAM, Walton E, Pingault JB, Provençal N, Pappa I, Vitaro F, et al. DRD4 methylation as a potential biomarker for physical aggression: an epigenome-wide, cross-tissue investigation. Am J Med Genet Part B Neuropsychiatr Genet. 2018. https://doi.org/10.1002/ajmg.b.32689.
Mitjans M, Seidel J, Begemann M, Bockhop F, Moya-Higueras J, Bansal V, et al. Violent aggression predicted by multiple pre-adult environmental hits. Mol Psychiatry. 2018. https://doi.org/10.1038/s41380-018-0043-3.
van Dongen J, Nivard MG, Baselmans BML, Zilhão NR, Ligthart L, Heijmans BT, et al. Epigenome-wide association study of aggressive behavior. Twin Res Hum Genet. 2015;18:686–98.
Lubke GH, McArtor DB, Boomsma DI, Bartels M. Genetic and environmental contributions to the development of childhood aggression. Dev Psychol. 2018;54:39–50.
Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266.
Jovanovic T, Vance LA, Cross D, Knight AK, Kilaru V, Michopoulos V, et al. Exposure to violence accelerates epigenetic aging in children. Sci Rep. 2017;7:1–7.
Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA, et al. Epigenetic aging in major depressive disorder. Am J Psychiatry. 2018:appi.ajp.2018.1.
Ori APS, Olde Loohuis LM, Guintivano J, Hannon E, Dempster E, St Clair D, et al. Schizophrenia is characterized by age- and sex-specific effects on epigenetic aging. bioRxiv. 2019:727859.
Thomas M, Achenbach CE. Manual for the child behavior checklist. Burlington 1991;7.
Goodman R. The Strengths and Difficulties Questionnaire: a research note. J Child Psychol Psychiatry. 1997;38:581–6.
Pulkkinen L, Kaprio J, Rose RJ. Peers, teachers and parents as assessors of the behavioural and emotional problems of twins and their adjustment: the Multidimensional Peer Nomination Inventory. Twin Res. 1999;2:274–85.
Achenbach TM, Rescorla LA. Manual for the ASEBA adult forms & profiles. English. 2003: University of Vermont, Research Center for Children.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders, (DSM IV). Washington DC, APA. 1994; Fourth Ed. 915.
Tellegen A, Lykken DT, Bouchard TJ, Wilcox KJ, Segal NL, Rich S. Personality similarity in twins reared apart and together. J Pers Soc Psychol. 1988;54:1031–9.
Wolf TM, Sklov MC, Wenzl PA, Hunter SM, Berenson GS. Validation of a measure of type A behavior pattern in children: Bogalusa heart study. Child Dev. 1982;53:126–35.
Ravaja N, Keltikangas-Järvinen L, Keskivaara P. Type A factors as predictors of changes in the metabolic syndrome precursors in adolescents and young adults–a 3-year follow-up study. Health Psychol. 1996;15:18–29.
Chen Y, Lemire M, Choufani S, Butcher DT, Zanke BW, Gallinger S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium Human Methylation 450 microarray. Epigenetics. 2013;2294:203–9.
Iterson M Van, Zwet EW Van, Heijmans BT, et al. Controlling bias and inflation in association studies using the empirical null distribution. Genome Biol. 2017:1–13.
Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010. https://doi.org/10.1093/bioinformatics/btq340.
Li M, Zou D, Li Z, Gao R, Sang J, Zhang Y, et al. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019. https://doi.org/10.1093/nar/gky1027.
Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436–47.
von Rhein D, Mennes M, van Ewijk H, Groenman AP, Zwiers MP, Oosterlaan J, et al. The NeuroIMAGE study: a prospective phenotypic, cognitive, genetic and MRI study in children with attention-deficit/hyperactivity disorder. Design and descriptives. Eur Child Adolesc Psychiatry. 2015. https://doi.org/10.1007/s00787-014-0573-4.
Freitag CM, Konrad K, Stadler C, De Brito SA, Popma A, Herpertz SC, et al. Conduct disorder in adolescent females: current state of research and study design of the FemNAT-CD consortium. Eur Child Adolesc Psychiatry. 2018;27:1077–93.
Hagenbeek FA, Roetman PJ, Pool R, Kluft C, Harms AC, van Dongen J, et al. Urinary amine and organic acid metabolites evaluated as markers for childhood aggression: the ACTION Biomarker study. Front Psychiatry. 2020;11:165.
van Dongen J, Nivard MG, Willemsen G, Hottenga J-J, Helmer Q, Dolan CV, et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016;7:11115.
Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–32.
Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019. https://doi.org/10.1038/s41588-018-0309-3.
Su KY, Li MC, Lee NW, Ho BC, Cheng CL, Chuang YC, et al. Perinatal polychlorinated biphenyls and polychlorinated dibenzofurans exposure are associated with DNA methylation changes lasting to early adulthood: Findings from Yucheng second generation. Environ Res. 2019. https://doi.org/10.1016/j.envres.2019.01.001.
Juricek L, Coumoul X. The aryl hydrocarbon receptor and the nervous system. Int J Mol Sci. 2018.
Chepelev NL, Moffat ID, Bowers WJ, Yauk CL. Neurotoxicity may be an overlooked consequence of benzo[a]pyrene exposure that is relevant to human health risk assessment. Rev Mutat Res. 2015;764:64–89.
Niu Q, Zhang H, Li X, Li M. Benzo[a]pyrene-induced neurobehavioral function and neurotransmitter alterations in coke oven workers. Occup Environ Med. 2010;67:444–8.
Malanchini M, Smith-Woolley E, Ayorech Z, Rimfeld K, Krapohl E, Vuoksimaa E, et al. Aggressive behaviour in childhood and adolescence: the role of smoking during pregnancy, evidence from four twin cohorts in the EU-ACTION consortium. Psychol Med. 2019. https://doi.org/10.1017/S0033291718001344.
Richardson TG, Richmond RC, North TL, Hemani G, Davey Smith G, Sharp GC, et al. An integrative approach to detect epigenetic mechanisms that putatively mediate the influence of lifestyle exposures on disease susceptibility. Int J Epidemiol. 2019;48:887–98.
Wiklund P, Karhunen V, Richmond R, Parmar P, Rodriguez A, De Silva M. DNA methylation links prenatal smoking exposure to later life health outcomes in offspring. Clin Epigenetics. 2019;11:97.
Odintsova VV, Roetman PJ, Ip HF, Pool R, Van der Laan CM, Tona DK, et al. Genomics of human aggression: current state of genome-wide studies and an automated systematic review tool. Psychiatr Genet. 2019;29:170–90.
Tielbeek JJ, Johansson A, Polderman TJC, Rautiainen M-R, Jansen P, Taylor M, et al. Genome-wide association studies of a broad spectrum of antisocial behavior. JAMA Psychiatry. 2017;74:1242.
Ip HF, van der Laan CM, Brikell I, Sánchez-Mora C, Nolte IM, St Pourcain B, et al. Genetic Association Study of Childhood Aggression across raters, instruments and age. bioRxiv. 2019:854927.
Relton CL, Gaunt T, McArdle W, Ho K, Duggirala A, Shihab H, et al. Data resource profile: Accessible Resource for Integrated Epigenomic Studies (ARIES). Int J Epidemiol. 2015. https://doi.org/10.1093/ije/dyv072.
Poulton R, Moffitt TE, Silva PA. The Dunedin Multidisciplinary Health and Development Study: overview of the first 40 years, with an eye to the future. Soc Psychiatry Psychiatr Epidemiol. 2015;50:679–93.
Moffitt TE, Adlam A, Affleck G, Andreou P, Aquan-Assee J, Arseneault L, et al. Teen-aged mothers in contemporary Britain. J Child Psychol Psychiatry Allied Discip. 2002;43:727–42.
Kaprio J. The Finnish Twin Cohort Study: an update. Twin Res Hum Genet. 2013;16:157–62.
Smith BH, Campbell A, Linksted P, Fitzpatrick B, Jackson C, Kerr SM, et al. Cohort profile: generation scotland: Scottish family health study (GS: SFHS). The study, its participants and their potential for genetic research on health and illness. Int J Epidemiol. 2013. https://doi.org/10.1093/ije/dys084.
Goldberg DP, Hillier VF. A scaled version of the General Health Questionnaire. Psychol Med. 1979. https://doi.org/10.1017/S0033291700021644.
Strandberg TE, Järvenpää AL, Vanhanen H, McKeigue PM. Birth outcome in relation to licorice consumption during pregnancy. Am J Epidemiol. 2001. https://doi.org/10.1093/aje/153.11.1085.
Vrijheid M, Slama R, Robinson O, Chatzi L, Coen M, van den Hazel P, et al. The human early-life exposome (HELIX): Project rationale and design. Environ Health Perspect. 2014;122:535–44.
Tigchelaar EF, Zhernakova A, Dekens JAM, Hermes G, Baranska A, Mujagic Z, et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772.
Rantakallio P. The longitudinal study of the northern Finland birth cohort of 1966. Paediatr Perinat Epidemiol. 1988;2:59–88.
Boomsma DI, Geus EJC, de, Vink JM, Stubbe JH, Distel MA, Hottenga J-J, et al. Netherlands twin register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–57.
Pedersen NL, McClearn GE, Plomin R, Nesselroade JR, Berg S, DeFaire U. The Swedish adoption twin study of aging: an update. Acta Genet Med Gemellol. 1991;40:7–20.
Buss AH, Plomin R. Temperament early developing personality traitsle. Hillsdale, NJ: Lawrence Erlbaum Associates Inc; 1984.
Plomin R, Pedersen NL, McClearn GE, Nesselroade JR, Bergeman CS. EAS temperaments during the last half of the life span: twins reared apart and twins reared together. Psychol Aging. 1988;3:43–50.
Raitakari OT, Juonala M, Rönnemaa T, Keltikangas-Järvinen L, Räsänen L, Pietikäinen M, et al. Cohort profile: the cardiovascular risk in young Finns study. Int J Epidemiol. 2008;37:1220–6.
L’Abée C, Sauer PJJ, Damen M, Rake JP, Cats H, Stolk RP. Cohort profile: the GECKO Drenthe study, overweight programming during early childhood. Int J Epidemiol. 2008;37:486–9.
Kruithof CJ, Kooijman MN, van Duijn CM, Franco OH, de Jongste JC, Klaver CCW, et al. The generation R study: biobank update 2015. Eur J Epidemiol. 2014;29:911–27.
Guxens M, Ballester F, Espada M, Fernández MF, Grimalt JO, Ibarluzea J, et al. Cohort profile: the INMA-INfancia y Medio Ambiente-(environment and childhood) project. Int J Epidemiol. 2012;41:930–40.
Witt SH, Frank J, Gilles M, Lang M, Treutlein J, Streit F, et al. Impact on birth weight of maternal smoking throughout pregnancy mediated by DNA methylation. BMC Genomics. 2018. https://doi.org/10.1186/s12864-018-4652-7.
Acknowledgements
This work was supported by ACTION. ACTION receives funding from the European Union Seventh Framework Program (FP7/2007–2013) under grant agreement no 602768. Cohort-specific acknowledgements are provided in eAppendix 1.
BIOS Consortium
Management team: Bastiaan T. Heijmans68, Peter A. C. ’t Hoen69, Joyce van Meurs70, Rick Jansen26, Lude Franke23.
Cohort collection: Dorret I. Boomsma1, René Pool1, Jenny van Dongen1, Jouke J. Hottenga1, Marleen M. J van Greevenbroek72, Coen D. A. Stehouwer72, Carla J. H. van der Kallen72, Casper G. Schalkwijk72, Cisca Wijmenga23, Lude Franke23, Sasha Zhernakova23, Ettje F. Tigchelaar23, P. Eline Slagboom68, Marian Beekman68, Joris Deelen68, Diana van Heemst73, Jan H. Veldink74, Leonard H. van den Berg74, Cornelia M. van Duijn71, Bert A. Hofman75, Aaron Isaacs71, André G. Uitterlinden70.
Data generation: Joyce van Meurs70, P. Mila Jhamai70, Michael Verbiest70, H. Eka D. Suchiman68, Marijn Verkerk70, Ruud van der Breggen68, Jeroen van Rooij70, Nico Lakenberg68.
Data management and computational infrastructure: Hailiang Mei76, Maarten van Iterson68, Michiel van Galen69, Jan Bot77, Dasha V. Zhernakova23, Rick Jansen26, Peter van ’t Hof76, Patrick Deelen23, Irene Nooren77, Peter A. C. ’t Hoen69, Bastiaan T. Heijmans68, Matthijs Moed68.
Data Analysis Group: Lude Franke23, Martijn Vermaat69, Dasha V. Zhernakova23, René Luijk68, Marc Jan Bonder23, Maarten van Iterson68, Patrick Deelen23, Freerk van Dijk78, Michiel van Galen69, Wibowo Arindrarto76, Szymon M. Kielbasa79, Morris A. Swertz78, Erik. W van Zwet79, Rick Jansen26, Peter-Bram ’t Hoen69, Bastiaan T. Heijmans68.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The following authors declare a conflict of interest: BF received educational speaking fees from Medice. AMM has received research support from Eli Lilly, Janssen, and The Sackler Trust and speaker fees from Illumina and Janssen. CMF has received funding by the DFG, BMBF, State of Hessen, and the EU. She receives royalties for books on ASD, ADHD, and MDD. The other authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Dongen, J., Hagenbeek, F.A., Suderman, M. et al. DNA methylation signatures of aggression and closely related constructs: A meta-analysis of epigenome-wide studies across the lifespan. Mol Psychiatry 26, 2148–2162 (2021). https://doi.org/10.1038/s41380-020-00987-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-00987-x
This article is cited by
-
Systemic interindividual epigenetic variation in humans is associated with transposable elements and under strong genetic control
Genome Biology (2023)
-
Integrative Multi-omics Analysis of Childhood Aggressive Behavior
Behavior Genetics (2023)
-
Trans-ancestry epigenome-wide association meta-analysis of DNA methylation with lifetime cannabis use
Molecular Psychiatry (2023)
-
DNA methylation in peripheral tissues and left-handedness
Scientific Reports (2022)
-
Associations of age, sex, sexual abuse, and genotype with monoamine oxidase a gene methylation
Journal of Neural Transmission (2021)