Abstract
How bone marrow niches regulate leukemogenic activities of leukemia-initiating cells (LICs) is unclear. The present study revealed that the metabolic niche component, ATP, efficiently induced ion influx in LICs through its ligand-gated ion channel, P2X1. P2X1 deletion impaired LIC self-renewal capacities and resulted in an approximately 8-fold decrease in functional LIC numbers in a murine acute myeloid leukemia (AML) model without affecting normal hematopoiesis. P2X1 phosphorylation at specific sites of S387 and T389 was essential for sustaining its promoting effects on leukemia development. ATP-P2X1-mediated signaling upregulated the PBX3 level to transactivate BCAT1 to maintain LIC fates. P2X1 knockdown inhibited the proliferation of both human AML cell lines and primary cells. The P2X1 antagonist sufficiently suppressed AML cell proliferation. These results provided a unique perspective on how metabolic niche factor ATP fine-tunes LIC activities, which may benefit the development of strategies for targeting LICs or other cancer stem cells.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is the most severe hematological malignancy and most commonly found in adults with high recurrence rates and mortality [1]. Leukemia-initiating cells (LICs) are a small cell population that is responsible for the initiation, progression, relapse and drug resistance of AML [2]. Although many intrinsic factors (i.e., transcription factors and epigenetic regulators) [3, 4] and extrinsic factors (bone marrow niche components) [5, 6] are required for LIC activities, there is still a lack of ideal targets for the eradication of LICs.
Increasing evidence shows that bone marrow niches play important roles in AML development [7, 8]. Many types of bone marrow niche cells and their secretory components, including growth factors, cytokines and metabolic products, are involved in LIC fate determinations [9, 10]. We recently also demonstrated that certain lipid metabolism regulators, such as ANGPTL2 and APOE, bind immune inhibitory receptors of LILRB2 and LILRB4, respectively, to significantly enhance AML-LIC self-renewal, migration or T cell suppression [11, 12]. These findings indicate that bone marrow metabolic niches may be tightly connected with LIC activities.
Adenosine triphosphate (ATP) mainly exists in cells and serves as a main energy carrier to support their biological behaviors. Various types of cells (e.g., neurons, platelets, lymphocytes and endothelial cells) also release ATP into the extracellular environment and contribute to many physiological and pathological activities [13, 14]. The extracellular ATP amounts in some solid tumors are increased to micromolar levels, and they promote tumorigenesis and chemoresistance by binding to purinergic receptors (P2Rs), including ligand-gated ion channels (P2Xs) and G protein-coupled membrane receptors (P2Ys) [15, 16]. P2X receptors (P2Xs) are cation-selective channels permeable to Na+, K+ or Ca2+, and ATP is the only known ligand for P2Xs, which are widely expressed in solid and hematological cancers [17, 18]. P2Xs contain seven family members (P2X1-7) with distinct ion channel properties and functions in response to ATP stimulation. For example, P2X1-4 can sensitively respond to extracellular ATP at nanomolar or micromolar levels, while P2X7 only responds to ATP at millimolar levels [19].
P2Xs have multifaceted roles in normal hematopoiesis or hematopoietic disorders. For example, P2X1 and P2X4 may be involved in the regulation of several fates of hematopoietic stem cells (HSCs) [20, 21]. P2X5 may be expressed in lymphoid malignancies, which can lead to LRH-1-specific cytotoxic T cell-mediated lysis [22]. P2X1- and P2X7-mediated signaling indirectly enhances the release of inflammatory factors from mesenchymal stem cells to regulate immune responses [23, 24]. P2X7 promotes AML development and impairs normal hematopoiesis via PBX3 signaling [25].
P2X1 is a nonselective ATP-gated and rapidly desensitizing ion channel expressed on smooth muscle cells [26], platelets [27], thymocytes [28] and neutrophils [29]. P2X1-mediated ion flux can be elicited at relatively low ATP levels (<10 nM), reach a peak within several seconds and last for more than 30 s. In contrast, high ATP levels trigger much faster ion flux than low ATP levels, reaching a peak within tens of milliseconds, and they are completely desensitized within seconds and quickly recover after desensitization [30, 31]. P2X1 plays important roles in male fertility, bladder contraction and platelet aggregation [32].
We recently demonstrated that ATP-P2X7-mediated signaling enhances AML development via the CREB-PHGDH pathway. Moreover, we noticed that several P2X members, except P2X7, such as P2X1, 4 and 5, were highly expressed in human AML samples, and P2X1 ranked top among them [33]. However, the detailed functions of the remaining P2X members, including P2X1, P2X4 and P2X5, in leukemogenesis remain largely unknown. Because P2X1 had the highest expression among all P2X members and was activated by extracellular ATP at the lowest level (nanomolar level), we evaluated its potential roles in sustaining LIC activities in the present study.
Results
P2X1 is highly expressed in LICs and required for their leukemogenic activities
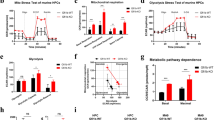
To evaluate the role of P2X1 in AML development, we first measured P2x1 mRNA levels in immunophenotypic Lin–Sca-1+c-Kit+CD34-Flt3- LT-HSCs and Mac-1+c-Kit+ LICs or Lin–Sca-1+c-Kit+CD34+CD16/CD32+ L-GMP cells (a population more highly enriched in LICs, Fig. 1A). We further demonstrated that immunophenotypic LICs or L-GMPs had approximately 20-fold higher expression levels of P2x1 than total AML cells, normal BM cells or HSCs (Fig. 1A), which was indicating that P2X1 may play key roles in leukemogenesis. Consistently, in vitro ATP simulation efficiently elicited ion flux in murine LICs, which was completely blocked by PPNDS, a selective P2X1 antagonist (Fig. 1B). To evaluate the potential roles of P2X1 in AML development, we knocked down P2x1 in murine AML cells using shRNAs (Fig. S1A), and demonstrated that leukemia development was significantly delayed in the recipient mice receiving P2x1-knockdown AML cells than WT counterparts as revealed by reduced leukemia cell frequencies in the peripheral blood (Figs. S1B, C) and prolonged overall survival of leukemic mice (47 or 56 vs. 36 days, Fig. S1D).
A The mRNA levels of P2x1 in normal mouse bone marrow cells (Normal-BM), Lin-Sca-1+c-Kit+CD34+Flk2+ ST-HSCs, Lin-Sca-1+c-Kit+CD34-Flk2- LT-HSCs, YFP+ leukemia cells (AML-BM), Mac-1+c-Kit+ LICs and Lin-Sca-1-c-Kit+CD34+CD16/32- L-GMP cells were measured by qRT–PCR (n = 3). B P2X1-mediated ion influx in murine Mac-1+c-Kit+ LICs was measured after sequential treatments with extracellular ATP and the PPNDS P2X1 antagonist by whole-cell patch-clamp recording. Data from three independent experiments. C Details of the nucleotide base sequences deleted in P2x1 knockout (P2x1-KO or P2x1-null) mice using the CRISPR-Cas9 strategy. D P2X1 protein levels in WT (WT #1-#2) and P2x1-KO (P2x1-KO #1-#2) BM cells were measured by western blot analysis. E Representative flow cytometric analysis of leukemia cells (YFP+) in the peripheral blood of recipient mice upon primary transplantation. F Quantification of the data shown in E (n = 5). G The overall survival of recipient mice transplanted with WT or P2x1-KO MLL-AF9+ BM cells upon primary transplantation (n = 5). H Representative images of the size of the spleens and livers of recipients upon primary transplantation. I Quantification of the data shown in H (n = 3). J Limiting dilution assays were performed with WT and KO BM YFP+ AML cells from the primary transplantation at the indicated cell doses, and functional LIC frequencies were calculated. Data are presented as the mean ± SD. One-way ANOVA with Tukey’s multiple comparison test (A), student’s two-tailed unpaired t test (F), log-rank test (G) and two-way ANOVA with Sidak’s multiple comparison test (I) were used for the comparison of statistical significance (*P < 0.05; **P < 0.01; and ***P < 0.001).
We then generated P2x1 knockout mice by the CRISPR-Cas9 technique with the deletion of 2 bp in exon 1 (Fig. 1C). P2X1 protein levels were undetected in total BM cells (Fig. 1D). We next established an MLL-AF9-induced murine AML model as previously described [34], and found that P2X1 deletion led to a notable delay in leukemia development in recipient mice receiving MLL-AF9-overexpressing P2x1-null Lin- BM cells as revealed by decreased YFP+ leukemic cell frequencies in the peripheral blood (4.3% vs. 21.4%, Fig. 1E, F and S1E, F), extended overall survival (91 vs. 62 days, Fig. 1G) and reduced leukemic cell infiltration in the spleens and livers of transplant mice (Fig. 1H, I and S1G). Limiting dilution assays with YFP+ AML cells from primary transplant recipients revealed that the frequency of functional WT LICs was 1 in 59, which was approximately 8.3-fold higher than that of P2x1-null control cells (1 in 491; Fig. 1J and Table S1).
In addition, we noticed that several P2X members, including P2X1, 4, 5 and 7, were highly expressed in AML cells. We also evaluated the roles of the remaining two P2X members in leukemogenesis, and demonstrated that P2X4 and P2X5 were not required for the maintenance of LIC leukemogenic activities (Fig. S1H, I), but P2X1 and P2X7 could enhance leukemogenic activities [33]. Interestingly, no significant difference in the percentages of LT-HSCs was observed between WT and P2x1-null mice (Fig. S1J, K). P2x1-null LT-HSCs also had repopulation abilities comparable to their WT counterparts (Fig. S1L–N). These data indicated that P2X1 supports leukemia development while sustaining normal hematopoiesis, which may be an ideal target for leukemia treatments.
P2X1 is important for the self-renewal of LICs
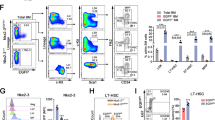
To further evaluate the role of P2X1 in sustaining LIC activities, we performed a serial transplantation with P2x1-null AML cells and revealed that the immunophenotypic Mac-1+c-Kit+ P2x1-null LIC frequency was notably decreased to 30.7% or 13.1% of WT LICs upon primary (Fig. 2A, B) or secondary transplantation (Fig. 2C), respectively. P2x1-null leukemic cells gave rise to significantly fewer colony numbers (Fig. 2D, E) and derived total cell counts (Fig. 2F) during the 1st and 2nd plating. Moreover, P2x1-null AML cells had impaired self-renewal abilities compared to WT controls as evidenced by decreased leukemic cell frequencies in the peripheral blood and significantly extended overall survival in the secondary transplantation assay (Fig. 2G, H). Consistently, the leukemic mice that received P2x1-knockdown AML cells had significantly longer overall survival than those that received scrambled cells (Fig. S2A, B). In addition, a significant difference in differentiation was observed after P2X1 deletion during the 1st and 2nd transplantation as measured by Mac-1/Gr-1 staining (Gr-1 level indicates the extent of differentiation, Fig. 2I–K). Meanwhile, we demonstrated that there was no difference in cell cycle between WT and P2x1-null LICs (Fig. S2C, D). However, there were much higher frequencies of early/late apoptotic cells in P2x1-null LICs compared to WT control cells (Fig. S2E, F). These results indicate that P2X1 may also promote AML development by suppressing LIC apoptosis.
A Representative flow cytometric analysis of Mac-1+c-Kit+ LICs in recipient mice upon primary transplantation. B, C The frequencies of WT and P2x1-KO Mac-1+c-Kit+ LICs from recipients upon primary (B) and secondary (C) transplantation (n = 3). D Representative images of colonies derived from WT and P2x1-KO YFP+ AML cells during the 1st and 2nd plating. E, F Colony numbers (E) and derived total cell counts (F) from WT and P2x1-KO YFP+ AML cells during the 1st and 2nd plating (n = 3). G Quantification of the frequencies of leukemia cells (YFP+) in the peripheral blood of recipient mice 3 weeks after transplantation (n = 5). H Overall survival was determined for the leukemic mice shown in G (n = 5). I Representative flow cytometric analysis of Mac-1+Gr1+ cells in recipient mice upon primary transplantation. J, K The frequencies of WT and P2x1-KO Mac-1+Gr1+ cells from the recipients upon primary (J) and secondary (K) transplantation (n = 3). Data are presented as the mean ± SD. Student’s two-tailed unpaired t test (B, C, G, J and K), log-rank test (H) and two-way ANOVA with Sidak’s multiple comparison test (E and F) were used for the comparison of statistical significance (**P < 0.01; and ***P < 0.001).
P2X1 regulates LIC activities via BCAT1-mediated branched chain amino acid metabolism
To understand the potential downstream targets of P2X1 in the regulation of LIC self-renewal capacities, we performed RNA-sequencing analysis of WT and P2x1-null immunophenotypic Mac-1+c-Kit+ LICs. Approximately 1383 upregulated and 864 downregulated genes were observed after P2X1 deletion (Fig. 3A). Among the top 10 significantly downregulated pathways according to the Kyoto Encyclopedia of Genes and Genome (KEGG) analyses, P2X1 was associated with the following pathways: valine, leucine and isoleucine metabolism (or branched chain amino acid (BCAA) metabolism); folate biosynthesis; and transcriptional misregulation in cancer (Fig. 3B). We then examined the expression levels of several key enzymes involved in BCAA catabolism (such as Bcat1, Bcat2, Bckdha, Bckdhb, Bckdk, Ppm1k and Dbt) and transcription factors (such as Meis1, Hoxa9, Ccnt1, Pbx1, Rora and Pbx3) that have been reported to be involved in the regulation of LIC activities from RNA-sequencing data. Interestingly, several genes, including Bcat2, Bckdhb, Bcat1, Hoxa9, Meis1, Ccnt1, Pbx3, Rora and Pbx1, were significantly decreased in P2x1-null LICs (Fig. S3A). Quantitative RT–PCR (qRT-PCR) further showed that Bcat1 (related to BCAA metabolism) or Hoxa9, Ccnt1, Pbx1, Rora and Pbx3 (related to transcriptional misregulation in cancer) were significantly reduced in P2x1-null LICs (Fig. 3C and Table S2), suggesting that these genes may act as downstream targets of P2X1.
A Volcano plots showing differentially expressed genes in WT and P2x1-KO Mac-1+c-Kit+ LICs. B KEGG pathway analysis was performed using mRNA-sequencing data from WT or P2x1-KO Mac-1+c-Kit+ LICs. C Potential candidates related to the “valine, leucine and isoleucine (branched chain amino acid, BCAA) metabolism” and “transcriptional misregulation in cancer” pathways were examined in WT and P2x1-KO Mac-1+c-Kit+ LICs by qRT–PCR (n = 3). D Representative flow cytometric analysis of YFP+mCherry+ leukemia cells (The markers for the indication of leukemia cells were YFP+ and mCherry+, which were the tags for the MSCV-MLL-AF9-YFP plasmid and MSCV-mCherry overexpression plasmid) in the peripheral blood of recipient mice transplanted with WT, P2x1-KO, Bcat1-overexpressing WT or P2x1-KO leukemia cells. E Quantification of the data shown in D (n = 5). F The overall survival was compared among the mice transplanted with WT, P2x1-KO, Bcat1-overexpressing WT or P2x1-KO leukemia cells (n = 5). G Representative images of the spleen and liver sizes of recipient mice transplanted with WT, P2x1-KO, Bcat1-overexpressing WT or P2x1-KO leukemia cells. H Quantification of the data shown in G (n = 3). I Representative images of colonies derived from WT, P2x1-KO, Bcat1-overexpressing WT or P2x1-KO YFP+mCherry+ AML cells. J, K Quantification of the colony numbers (J) and derived total cell counts (K) in I (n = 3). L The Bcat1 mRNA levels in BM cells from recipient mice transplanted with WT, P2x1-KO, Bcat1-overexpressing WT or P2x1-KO AML cells were measured by qRT–PCR (n = 3). Data are presented as the mean ± SD. One-way ANOVA with Tukey’s multiple comparison test (E, J, K and L), log-rank test (F) and two-way ANOVA with Sidak’s multiple comparison test (H) were used for the comparison of statistical significance (*P < 0.05; **P < 0.01; and ***P < 0.001).
BCAT1-mediated BCAA metabolism has been reported to be essential in sustaining LIC activities via enhanced mTOR signaling pathways [35]. PBX1 and PBX3 are two key transcription factors involved in the maintenance of stem cell activities and tumorigenesis [36, 37]. Therefore, we first overexpressed Bcat1 in P2x1-null AML cells and showed that ectopic overexpression of Bcat1 almost completely rescued the P2X1 loss of function in recipient mice as evidenced by increased leukemic cell frequencies in the peripheral blood (Fig. 3D, E), reduced overall survival (Fig. 3F) and enhanced infiltration in spleens and livers (Fig. 3G, H). Bcat1 overexpression had no effect on WT AML cell growth in vivo (Fig. 3D–H). An in vitro colony formation assay also revealed that overexpression of Bcat1 in P2x1-null AML cells resulted in increased colony numbers and total derived cell numbers (Fig. 3I–K). The mRNA expression levels of Bcat1 in WT and P2x1-null AML cells were validated by qRT–PCR (Fig. 3L). Consistently, ectopic overexpression of Bcat1 in P2x1 knockdown AML cells decreased the leukemia cell frequencies in the peripheral blood (Fig. S3B, C) and overall survival of recipient mice (Fig. S3D). We also measured the expression levels of LIC associated genes Meis1, Hoxa9, Pbx1 and Pbx3 in Mac-1+c-Kit+ LICs from recipient mice transplanted with WT, P2x1-null, Bcat1-overexpressing WT or P2x1-null AML cells by qRT-PCR. Interestingly, Bcat1 overexpression can restore the expression of Meis1 and Hoxa9, but not Pbx1 and Pbx3 (Fig. S3E).
To understand the potential roles of P2X1 in LIC activities, we further examined the potential candidates involved in LIC self-renewal and differentiation. Several genes that have been reported to be related to the self-renewal (Creb, Hoxb4, Hoxa9, Myb, Cbx5, Meis1, Bmi1, Hmgb3 and Pbx1) of AML cells were indeed decreased in the P2x1-null LICs, whereas the differentiation gene expression signatures (Gfi1b, Gata2, Cebpe and Cebpg) were increased (Fig. S3F) [4, 38,39,40,41]. Consistently, qRT-PCR further demonstrated that most of the self-renewal genes, including Stat5b, Creb, Hoxa9, Myb, Cbx5, Meis1, Bmi1, Humg3 and Pbx1, were significantly downregulated in the P2x1-null LICs. In contrast, the differentiation related genes, including Gfi1b, Gata2, Cebpg, Klf4 and Pu.1, were significantly upregulated in the P2x1-null LICs (Fig. S3G).
PBX3 transactivates BCAT1 expression to sustain LIC activities
To further delineate the upstream targets of BCAT1, we evaluated several potential signaling pathways downregulated in P2x1-null LICs, such as transcriptional misregulation in cancer, and we found that the Pbx3 mRNA level in P2x1-null LICs was significantly decreased to 10% of that in WT LICs (Fig. 3C). We then overexpressed Pbx3 in P2x1-null AML cells and demonstrated that its ectopic expression enhanced AML progression in recipient mice as indicated by increased leukemic cell frequencies in the peripheral blood (Fig. 4A, B), reduced overall survival (Fig. 4C, D) and enhanced infiltration in spleens and livers (Fig. S4A). Consistently, Pbx3-overexpressing P2x1-null AML cells had enhanced clonogenic capacities as well as increased colony numbers and total derived leukemic cells (Fig. S4B–D). We also measured the Bcat1 mRNA levels in WT, P2x1-null, Pbx3-overexpressing WT or P2x1-null leukemia cells and showed that the Bcat1 mRNA level was significantly downregulated in P2x1-null AML cells, while increased in Pbx3-overexpressing leukemic cells (Fig. S4E).
A Representative flow cytometric analysis of YFP+mCherry+ leukemia cells (The markers for the indication of leukemia cells were YFP+ and mCherry+, which were the tags for the MSCV-MLL-AF9-YFP plasmid and MSCV-mCherry overexpression plasmid) in the peripheral blood of recipient mice transplanted with WT, P2x1-KO, Pbx3-overexpressing WT or P2x1-KO leukemia cells. B Quantification data in A (n = 5). C The overall survival was compared among the mice transplanted with WT, P2x1-KO, Pbx3-overexpressing WT or P2x1-KO leukemia cells (n = 5). D The mRNA levels of Pbx3 in BM cells of recipient mice transplanted with WT, P2x1-KO, Pbx3-overexpressing WT or P2x1-KO AML cells were measured by qRT–PCR (n = 3). E A luciferase reporter assay was used to evaluate PBX3 transcriptional activities on Bcat1 (n = 3). Data from three independent experiments. F The binding of PBX3 to the Bcat1 promoter region was determined by a ChIP assay. Data are presented as the mean ± SD. One-way ANOVA with Tukey’s multiple comparison test (B and D) and log-rank test (C) were used for the comparison of statistical significance (*P < 0.05; **P < 0.01; and ***P < 0.001).
To determine potential connections between PBX3 and BCAT1, we performed a dual-luciferase reporter assay and found that PBX3 directly bound the Bcat1 promoter to transactivate its expression in a dose-dependent manner (Fig. 4E), which was further confirmed by a ChIP experiment (Fig. 4F). As mentioned above, BCAT1-mediated BCAA metabolism may be involved in the regulation of mTOR signaling and mitochondrial function, we then examined the mitochondrial metabolisms upon P2x1 deletion. Interestingly, P2X1 loss could cause the decrease in the mitochondrial membrane potential and ROS levels (Fig. S4F, G). Consistently, P2X1-null AML cells had much lower levels of glycolysis and oxidative phosphorylation as evidenced by the notable reduction in ATP, ECARs, and OCRs than WT counterparts (Fig. S4H–J).
P2X1 phosphorylation sites at S387 and T389 are critical for the leukemogenic activities of LICs
Because P2X1 can be phosphorylated at several phosphorylation sites [42], including S387, S388 and T389, we investigated whether these phosphorylation sites play a role in AML development by mutating the abovementioned P2X1 phosphorylation sites (Table S2) and evaluating their potential effects on leukemogenesis. Among the S387A, S388A and T389A mutants, neither S387A nor T389A rescued the impaired leukemogenic activities of the P2x1-knockdown murine AML cell line, C1498 (Fig. S5A, B). The colony formation assay showed that overexpression of both P2x1-S387A and P2x1-T389A in P2x1-null AML cells did not affect in vitro cell growth compared to P2x1-null control cells (Fig. 5A–C). In vivo transplantation experiments showed that the recipient mice receiving P2x1-null AML cells with ectopic expression of either P2x1-S387A or the P2x1-T389A mutant had similar overall survival to the mice receiving P2x1-null AML cells (Fig. 5D–F). The overexpression levels of P2x1-WT, P2x1-S387A and P2x1-T389A in P2x1-null AML were validated by qRT–PCR (Fig. 5G). In contrast, overexpression of the P2x1-S388A mutant and P2x1-WT control in P2x1-null AML cells had a similar effect on cell proliferation as indicated by the in vitro colony formation assay (Fig. S5C–E) and the in vivo transplantation assay (Fig. S5F–H).
A Representative images of colonies derived from WT and P2x1-KO AML cells ectopically expressing empty vector (EV), WT or mutant P2x1 (S387A or T389A). B, C Quantification of the colony numbers (B) and derived total cell counts (C) shown in A (n = 3). D Representative flow cytometric analysis of YFP+mCherry+ leukemia cells (The markers for the indication of leukemia cells were YFP+ and mCherry+, which were the tags for the MSCV-MLL-AF9-YFP plasmid and MSCV-mCherry overexpression plasmid) in the peripheral blood of recipient mice transplanted with WT, P2x1-KO AML cells ectopically expressing empty vector (EV), WT or mutant P2x1 (S387A or T389A). E Quantification of data shown in D. One representative out of two independent experiments with n = 5 mice per group is shown. F The overall survival was compared among the mice transplanted with WT, P2x1-KO AML cells ectopically expressing empty vector (EV), WT or mutant P2x1 (S387A or T389A) (n = 5). G The P2x1 mRNA levels in BM cells of recipient mice transplanted with WT, P2x1-KO AML cells ectopically expressing empty vector (EV), WT or mutant P2x1 (S387A or T389A) were measured by qRT–PCR (n = 3). Data are presented as the mean ± SD. One-way ANOVA with Tukey’s multiple comparison test (B, C, E and G) and log-rank test (F) were used for the comparison of statistical significance (*P < 0.05; **P < 0.01; and ***P < 0.001).
P2X1 is required for the proliferation of human AML cells
We have previously reported that P2X1 has the highest mRNA expression levels in AML cells among all P2X members according to TCGA databases [33]. We analyzed TCGA data and showed that P2X1 mRNA levels in human AML cells were significantly higher than those in normal BM cells (Fig. 6A), which were adversely correlated with patient overall survival (TCGA and http://www.genomicscape.com) (Fig. S6A and Fig. 6B). P2X1 was highly expressed in several human AML cell lines, especially in FAB-M5-type AML cells, including THP-1 (M5), U937 (M5) and MV4-11 (M5) cells (Fig. S6B). AML cell lines from other FAB AML types, such as HL60 (M3) and NB4 (M3) cells, had relatively low P2X1 mRNA levels. We knocked down P2X1 in these cell lines using two shRNAs (Fig. S6C) and demonstrated that P2X1 deficiency resulted in significantly delayed in vitro cell growth in THP-1, U937 and MV4-11 cells (Fig. S6D–F). An in vivo transplantation assay showed that the recipient mice receiving P2X1-knockdown THP-1 cells had reduced leukemia cell frequencies in the peripheral blood (Fig. S6G) and prolonged overall survival (24 or 38 days vs. 15 days, Fig. S6H) compared to control mice.
A The P2X1 mRNA levels were analyzed in AML cells and normal BM cells from TCGA databases (AML, n = 173; normal, n = 70). B Relationship between P2X1 mRNA levels and overall survival in AML patients from the Genomicscape dataset. C The mRNA levels of P2X1 in human normal cord blood cells (Normal-total), normal CD34- cells (Normal-non-HSCs), normal Lin-CD34+CD38-CD45RA-CD90+ HSCs (Normal-HSCs), human primary AML cells (AML-total), CD34- cells (AML-non-LICs) and human primary CD34+ AML-LICs (AML-LICs) were measured by qRT–PCR (n = 3). D The P2X1-mediated ion influx in human CD34+ LICs was measured after sequential treatments with extracellular ATP and the PPNDS P2X1 antagonist by whole-cell patch-clamp recording (n = 3). E–G The numbers of human primary AML cells were counted at the indicated days upon P2X1 knockdown by shRNAs (sh-P2X1 #1 and #2) and a scrambled control. Three patient samples were used for the indicated experiments (AML #1-#3) (n = 3). (H) Representative images of colonies derived from human AML cells after P2X1 knockdown by shRNAs (sh-P2X1 #1 and #2) or a scrambled control. I, J Colony numbers (I) and derived total cell counts (J) were counted in human AML cells after P2X1 knockdown by shRNAs (sh-P2X1 #1 and #2) and scrambled controls (n = 3). Data are presented as the mean ± SD. Student’s two-tailed unpaired t test (A), one-way ANOVA with Tukey’s multiple comparison test (C) and two-way ANOVA with Sidak’s multiple comparison test (E–G, I and J) were used for the comparison of statistical significance (*P < 0.05; **P < 0.01; and ***P < 0.001).
In the present study, we also found human AML-LICs had the highest expression levels of P2X1 among non-LICs, total AML cells, normal HSCs, normal-non-HSCs and normal total hematopoietic cells (Fig. 6C). A whole-cell patch-clamp recording assay showed that P2X1-mediated ion flux was elicited upon ATP stimulation and selectively inhibited by PPNDS (Fig. 6D). In addition, P2X1-knockdown immunophenotypic human primary CD34+ AML-LICs grew much slower in vitro (Fig. 6E–G) and resulted in approximately 50% fewer colony counts or 40% fewer derived total AML cells than the control cells (Fig. 6H–J).
Mechanistically, we noticed that knockdown of P2X1 in either human primary AML-LICs (Fig. S6I–K) or human AML cell lines (THP-1, U937, MV4-11; Fig. S6L–N) resulted in the significant reduction in the expression levels of BCAT2, BCAT1, MEIS1, HOXA9, PBX1, and PBX3. Furthermore, we had mutated the phosphorylation sites in human AML cell lines and found that neither S387 A nor T389A rescued the impaired leukemogenic activities of the P2X1-knockdown human AML cell lines, such as THP-1 and MV4-11 cells (Fig. S6O, P). These data suggested that P2X1 is important for the leukemogenic activities of both human AML cell lines and primary AML-LICs.
P2X1 antagonist efficiently inhibits AML cell proliferation
To evaluate potential applications of P2X1 blockade in AML treatment, we examined the blocking effect of the P2X1 antagonist, PPNDS, on the ion flux triggered by extracellular ATP. The results showed that PPNDS selectively inhibited ion flux in THP-1 cells (Fig. 7A). Both murine (C1498) and human (THP-1) AML cell lines grew significantly slower than control cells in a dose-dependent manner after PPNDS treatment (Fig. 7B, C). In contrast, PPNDS did not affect the proliferation of normal hematopoietic cells, such as human cord blood CD34+ hematopoietic stem progenitor cells (Fig. 7D). Because no suitable P2X1 antagonists were available for in vivo treatment in leukemic mice, we performed an in vitro functional colony formation assay. PPNDS treatment resulted in an approximately 30% decrease in colony numbers and total derived cell counts from murine WT AML cells but not P2x1-null AML cells (Fig. 7E–G). Consistently, PPNDS sufficiently suppressed the colony formation of human primary AML cells as indicated by a reduction in colony numbers and their derived cell numbers (Fig. 7H, I), suggesting that PPNDS may provide a novel way to efficiently target AML cells or LICs in the clinic. In summary, ATP-P2X1-mediated signaling was required for the leukemogenic activities of both murine and human LICs. P2X1 phosphorylation levels at specific sites and the downstream PBX3-BCAT1 pathways enhanced AML development (Fig. 7J).
A The P2X1-mediated ion influx in THP-1 cells was measured upon sequential treatments with extracellular ATP and the P2X1 antagonist PPNDS by whole-cell patch-clamp recording. Data from three independent experiments. B–D The indicated doses of PPNDS were used for in vitro treatments of C1498 cells (B), THP-1 cells (C) and human cord blood CD34+ hematopoietic stem progenitor cells (D) followed by the determination of total cell counts (n = 3). E Representative images of colonies derived from WT and P2x1-KO murine AML cells treated with PPNDS (5 μM) or PBS. F, G The colony numbers (F) and derived total cell counts (G) of WT and P2x1-KO murine AML cells were counted after treatment with PPNDS (5 μM) or PBS for 5 days (n = 3). H, I The colony numbers (H) and derived total cell counts (I) of patient primary AML cells were counted after treatment with PPNDS (5 μM) or PBS for 7 days (n = 3). J A working model for the effect of P2X1 on the fate determination of AML-LICs, including self-renewal and differentiation, which is fine-tuned by P2X1 phosphorylation levels at sites of S387 or T389 and its downstream PBX3-BCAT1 pathways (S serine; T threonine; A alanine). Data are presented as the mean ± SD. Two-way ANOVA with Sidak’s multiple comparison test (F–I) were used for the comparison of statistical significance (*P < 0.05; **P < 0.01; and ***P < 0.001).
Discussion
The mechanism by which microenvironmental/niche components contribute to tumorigenesis or leukemogenesis is poorly understood. Extracellular ATP amounts in niches are relatively low (at nanomolar levels) in normal tissues, but they can be increased up to micromolar levels in many pathological states [15, 43]. Although it has been reported that extracellular ATP levels enhance the proliferation of cancer cells [44, 45], some studies have also shown that extracellular ATP inhibits cancer cell growth [46]. These controversial findings may be due to the differential expression of different P2 receptor subtypes [47]. Our previous study revealed that P2X7 enhance leukemia progress via PHGDH-mediated serine metabolic pathways [33]. In the present study, we further showed that P2X1, but not P2X4 or P2X5, promoted leukemogenesis through the PBX3-BCAT1 pathways. The mechanisms by which only P2X1 and P2X7 are involved in AML development, but not P2X4 and P2X5 (Fig. S1H, I), remain unclear, although all four members are highly expressed in AML cells [33, 48].
In the present study, we demonstrated that P2X1 served as an oncogene to enhance LIC self-renewal. However, the roles of P2X1 in other AML types or other leukemia types remain to be further investigated. Interestingly, we showed that P2X1 knockdown in several human primary leukemia cell lines from other FAB-AML types also led to a significant delay in cell proliferation, indicating that P2X1 may exert its oncogenic effects on a variety of cancer types. P2X1 has been reported to enhance mitochondrial activities in T-cell acute lymphoblastic leukemia (T-ALL) cells [49]. Consistently, we also showed that P2X1 augmented BCAT1-mediated BCAA catabolism, which is reported to contribute to enhanced oxidative phosphorylation [50, 51]. These results implicate that metabolic niche factors, such as ATP, may be tightly connected with the intrinsic metabolism of leukemia cells.
P2X1 has been reported to be basally phosphorylated at multiple sites, and it has a conserved intracellular protein kinase C (PKC) motif of TXK/R at the amino terminus [42, 52]. Phosphorylation at the T18P19R20 PKC sites of P2X1 may be required for its physiological functions, and mutation of these proteins leads to an increase in the desensitization rate and a decrease in the peak amplitude of ion flux [52]. However, P2X1 activities may not be directly induced by its phosphorylation level but may be mediated by staurosporine-sensitive phosphorylation of its cofactors [53]. Interestingly, we showed that P2X1 phosphorylation at sites S387 and T389, but not S388, was critical for its effects on enhancing AML development. However, it remains unclear how these two functional phosphorylation sites are regulated. Moreover, the underlying mechanisms related to P2X1 phosphorylation levels in the upregulation of PBX3 expression remain unclear.
Although relapse and drug resistance commonly occur, chemotherapy is still the major treatment for AML in the clinic to date. We have previously shown that inhibition of P2X7 by its specific antagonist (A-740003) efficiently delays AML development [33]. In the present study, we also showed that the P2X1 antagonist (PPNDS) significantly decreased AML cell proliferation. It is possible that combinational treatments targeting both P2X1 and P2X7 may be more potent in blocking AML progression or drug resistance. More efforts are required for screening other antagonists with better performance in AML treatment. In summary, the present study demonstrated that P2X1 enhances LIC self-renewal and suppresses its differentiation via PBX3-BCAT1 signaling without affecting normal hematopoiesis. These results indicated that targeting P2X1 may be an efficient approach for the treatment of AML or other cancers.
Methods
Animals
The CD45.1 mice were provided by Dr. Jiang Zhu at Ruijin Hospital, Shanghai, China. The P2x1-knockout (KO), P2x4-knockout (P2x4-KO) and P2x5-knockout (P2x5-KO) mice in a C57BL/6 background were generated at the Animal Core Facility at the School of Basic Medicine, Shanghai Jiao Tong University School of Medicine. C57BL/6 CD45.2 and NOD-SCID mice were ordered from Shanghai SLAC Laboratory Animal Co. Ltd and maintained at Animal Core Facility at Shanghai Jiao Tong University School of Medicine. All animal experiments were approved and conducted according to the Guidelines for Animal Care at Shanghai Jiao Tong University School of Medicine. All the other information related to the Methods is in the Supplementary Data file.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. Further inquiries can be directed to the corresponding authors.
References
Khwaja A, Bjorkholm M, Gale RE, Levine RL, Jordan CT, Ehninger G, et al. Acute myeloid leukaemia. Nat Rev Dis Prim. 2016;2:16010.
Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature 2008;453:1072–8.
Wong SH, Goode DL, Iwasaki M, Wei MC, Kuo HP, Zhu L, et al. The H3K4-Methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell. 2015;28:198–209.
Park SM, Cho H, Thornton AM, Barlowe TS, Chou T, Chhangawala S, et al. IKZF2 drives leukemia stem cell self-renewal and inhibits myeloid differentiation. Cell Stem Cell. 2019;24:153–65.
Krause DS, Fulzele K, Catic A, Sun CC, Dombkowski D, Hurley MP, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19:1513–7.
Agarwal P, Isringhausen S, Li H, Paterson AJ, He J, Gomariz A, et al. Mesenchymal niche-specific expression of Cxcl12 controls quiescence of treatment-resistant leukemia stem cells. Cell Stem Cell. 2019;24:769–84.
Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114:1150–7.
Ghobrial IM, Detappe A, Anderson KC, Steensma DP. The bone-marrow niche in MDS and MGUS: implications for AML and MM. Nat Rev Clin Oncol. 2018;15:219–33.
Baryawno N, Przybylski D, Kowalczyk MS, Kfoury Y, Severe N, Gustafsson K, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–32.
Forte D, Garcia-Fernandez M, Sanchez-Aguilera A, Stavropoulou V, Fielding C, Martin-Perez D, et al. Bone marrow mesenchymal stem cells support acute myeloid leukemia bioenergetics and enhance antioxidant defense and escape from chemotherapy. Cell Metab. 2020;32:829–43.
Zheng J, Umikawa M, Cui C, Li J, Chen X, Zhang C, et al. Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature. 2012;485:656–60.
Deng M, Gui X, Kim J, Xie L, Chen W, Li Z, et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature. 2018;562:605–9.
Faas MM, Saez T, de Vos P. Extracellular ATP and adenosine: The Yin and Yang in immune responses? Mol Asp Med. 2017;55:9–19.
Chiarella AM, Ryu YK, Manji GA, Rustgi AK. Extracellular ATP and adenosine in cancer pathogenesis and treatment. Trends Cancer. 2021;7:731–50.
Di Virgilio F, Sarti AC, Falzoni S, De Marchi E, Adinolfi E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat Rev Cancer. 2018;18:601–18.
Mimoto F, Tatsumi K, Shimizu S, Kadono S, Haraya K, Nagayasu M, et al. Exploitation of elevated extracellular ATP to specifically direct antibody to tumor microenvironment. Cell Rep. 2020;33:108542.
Adinolfi E, Melchiorri L, Falzoni S, Chiozzi P, Morelli A, Tieghi A, et al. P2X7 receptor expression in evolutive and indolent forms of chronic B lymphocytic leukemia. Blood. 2002;99:706–8.
Adinolfi E, Raffaghello L, Giuliani AL, Cavazzini L, Capece M, Chiozzi P, et al. Expression of P2X7 receptor increases in vivo tumor growth. Cancer Res. 2012;72:2957–69.
North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–67.
Casati A, Frascoli M, Traggiai E, Proietti M, Schenk U, Grassi F. Cell-autonomous regulation of hematopoietic stem cell cycling activity by ATP. Cell Death Differ. 2011;18:396–404.
Adamiak M, Bujko K, Thapa A, Pensato V, Brzezniakiewicz-Janus K, Ratajczak J, et al. The P2X4 purinergic receptor has emerged as a potent regulator of hematopoietic stem/progenitor cell mobilization and homing-a novel view of P2X4 and P2X7 receptor interaction in orchestrating stem cell trafficking. Leukemia. 2022;36:248–56.
Overes IM, de Rijke B, van Horssen-Zoetbrood A, Fredrix H, de Graaf AO, Jansen JH, et al. Expression of P2X5 in lymphoid malignancies results in LRH-1-specific cytotoxic T-cell-mediated lysis. Br J Haematol. 2008;141:799–807.
Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature. 2014;509:310–7.
Jiang LH, Hao Y, Mousawi F, Peng H, Yang X. Expression of P2 purinergic receptors in mesenchymal stem cells and their roles in extracellular nucleotide regulation of cell functions. J Cell Physiol. 2017;232:287–97.
Feng W, Yang X, Wang L, Wang R, Yang F, Wang H, et al. P2X7 promotes the progression of MLL-AF9 induced acute myeloid leukemia by upregulation of Pbx3. Haematologica 2021;106:1278–89.
Ralevic V. P2X receptors in the cardiovascular system and their potential as therapeutic targets in disease. Curr Med Chem. 2015;22:851–65.
Mahaut-Smith MP, Jones S, Evans RJ. The P2X1 receptor and platelet function. Purinergic Signal. 2011;7:341–56.
Chvatchko Y, Valera S, Aubry JP, Renno T, Buell G, Bonnefoy JY. The involvement of an ATP-gated ion channel, P(2X1), in thymocyte apoptosis. Immunity. 1996;5:275–83.
Lecut C, Frederix K, Johnson DM, Deroanne C, Thiry M, Faccinetto C, et al. P2X1 ion channels promote neutrophil chemotaxis through Rho kinase activation. J Immunol. 2009;183:2801–9.
Rettinger J, Schmalzing G. Desensitization masks nanomolar potency of ATP for the P2X1 receptor. J Biol Chem. 2004;279:6426–33.
Lorinczi E, Bhargava Y, Marino SF, Taly A, Kaczmarek-Hajek K, Barrantes-Freer A, et al. Involvement of the cysteine-rich head domain in activation and desensitization of the P2X1 receptor. Proc Natl Acad Sci USA. 2012;109:11396–401.
Fong Z, Griffin CS, Large RJ, Hollywood MA, Thornbury KD, Sergeant GP. Regulation of P2X1 receptors by modulators of the cAMP effectors PKA and EPAC. Proc Natl Acad Sci USA. 2021;118:275–87.
He X, Wan J, Yang X, Zhang X, Huang D, Li X, et al. Bone marrow niche ATP levels determine leukemia-initiating cell activity via P2X7 in leukemic models. J Clin Invest. 2021;131:1–15.
Zhang Y, Xia F, Liu X, Yu Z, Xie L, Liu L, et al. JAM3 maintains leukemia-initiating cell self-renewal through LRP5/AKT/beta-catenin/CCND1 signaling. J Clin Invest. 2018;128:1737–51.
Gu Z, Liu Y, Cai F, Patrick M, Zmajkovic J, Cao H, et al. Loss of EZH2 reprograms BCAA metabolism to drive leukemic transformation. Cancer Disco. 2019;9:1228–47.
Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121:1422–31.
Pi WC, Wang J, Shimada M, Lin JW, Geng H, Lee YL, et al. E2A-PBX1 functions as a coactivator for RUNX1 in acute lymphoblastic leukemia. Blood. 2020;136:11–23.
Salik B, Yi H, Hassan N, Santiappillai N, Vick B, Connerty P, et al. Targeting RSPO3-LGR4 signaling for leukemia stem cell eradication in acute myeloid leukemia. Cancer Cell. 2020;38:263–78.
Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–40.
Li Y, He Y, Liang Z, Wang Y, Chen F, Djekidel MN, et al. Alterations of specific chromatin conformation affect ATRA-induced leukemia cell differentiation. Cell Death Dis. 2018;9:200.
Thivakaran A, Botezatu L, Hones JM, Schutte J, Vassen L, Al-Matary YS, et al. Gfi1b: a key player in the genesis and maintenance of acute myeloid leukemia and myelodysplastic syndrome. Haematologica. 2018;103:614–25.
Roberts JA, Bottrill AR, Mistry S, Evans RJ. Mass spectrometry analysis of human P2X1 receptors; insight into phosphorylation, modelling and conformational changes. J Neurochem. 2012;123:725–35.
Kepp O, Bezu L, Yamazaki T, Di Virgilio F, Smyth MJ, Kroemer G, et al. ATP and cancer immunosurveillance. EMBO J. 2021;40:e108130.
Li XY, Moesta AK, Xiao C, Nakamura K, Casey M, Zhang H, et al. Targeting CD39 in cancer reveals an extracellular ATP- and inflammasome-driven tumor immunity. Cancer Disco. 2019;9:1754–73.
Yang H, Geng YH, Wang P, Yang H, Zhou YT, Zhang HQ, et al. Extracellular ATP promotes breast cancer invasion and chemoresistance via SOX9 signaling. Oncogene. 2020;39:5795–810.
Adinolfi E, De Marchi E, Orioli E, Pegoraro A, Di Virgilio F. Role of the P2X7 receptor in tumor-associated inflammation. Curr Opin Pharm. 2019;47:59–64.
White N, Burnstock G. P2 receptors and cancer. Trends Pharm Sci. 2006;27:211–7.
Chen S, Feng W, Yang X, Yang W, Ru Y, Liao J, et al. Functional expression of P2X family receptors in macrophages is affected by microenvironment in mouse T cell acute lymphoblastic leukemia. Biochem Biophys Res Commun. 2014;446:1002–9.
Ledderose C, Woehrle T, Ledderose S, Strasser K, Seist R, Bao Y, et al. Cutting off the power: inhibition of leukemia cell growth by pausing basal ATP release and P2X receptor signaling? Purinergic Signal. 2016;12:439–51.
Li T, Zhang Z, Kolwicz SC Jr, Abell L, Roe ND, Kim M, et al. Defective branched-chain amino acid catabolism disrupts glucose metabolism and sensitizes the heart to ischemia-reperfusion injury. Cell Metab. 2017;25:374–85.
Zhang B, Xu F, Wang K, Liu M, Li J, Zhao Q, et al. BCAT1 knockdown-mediated suppression of melanoma cell proliferation and migration is associated with reduced oxidative phosphorylation. Am J Cancer Res. 2021;11:2670–83.
Liu GJ, Brockhausen J, Bennett MR. P2X1 receptor currents after disruption of the PKC site and its surroundings by dominant negative mutations in HEK293 cells. Auton Neurosci. 2003;108:12–6.
Vial C, Tobin AB, Evans RJ. G-protein-coupled receptor regulation of P2X1 receptors does not involve direct channel phosphorylation. Biochem J 2004;382:101–10.
Funding
This work was supported by grants from National Basic Research Program of China (2019YFA0801800 and 2018YFA0107000), National Natural Science Foundation of China (81825001, 32030030, 32030065, 31971052, 81900147, 82000147, 32100906, 82170175), the innovative group of NSFC (81721004), Shanghai Science and Technology Commission (19XD1422100, 20ZR1430900, 20JC1410100 and 20204Y0008), CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-051), the Fundamental Research Funds for the Central Universities, and Shanghai Frontiers Science Center of Cellular Homeostasis and Human Diseases.
Author information
Authors and Affiliations
Contributions
X.H., Y.X., C.C., J.W., Y.Z., and J.Z. designed the experiments, performed the experiments, analyzed data and wrote the paper; D.H., Z.Y., J.Y., L.X., L.L., and Y.Y. performed the experiments, provided reagents and helped with the manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
Human BM mononuclear cells of AML samples were kindly provided by the Department of Hematology at Xinhua Hospital, Shanghai Jiao Tong University School of Medicine. The patient’s written informed consent was obtained and approved by the Ethics Committee for Medical Research (IRB) of Shanghai Jiao Tong University School of Medicine.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, X., Xu, Y., Huang, D. et al. P2X1 enhances leukemogenesis through PBX3-BCAT1 pathways. Leukemia 37, 265–275 (2023). https://doi.org/10.1038/s41375-022-01759-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01759-y
This article is cited by
-
Purinergic pathways and their clinical use in the treatment of acute myeloid leukemia
Purinergic Signalling (2024)