Abstract
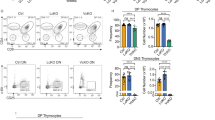
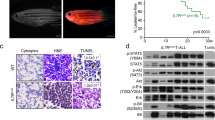
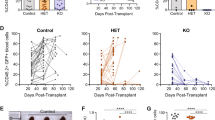
The majority of cases of T-cell acute lymphoblastic leukemia (T-ALL) contain chromosomal abnormalities that drive overexpression of oncogenic transcription factors. However, whether these initiating oncogenes are required for leukemia maintenance is poorly understood. To address this, we developed a tetracycline-regulated mouse model of T-ALL driven by the oncogenic transcription factor Lmo2. This revealed that whilst thymus-resident pre-Leukemic Stem Cells (pre-LSCs) required continuous Lmo2 expression, the majority of leukemias relapsed despite Lmo2 withdrawal. Relapse was associated with a mature phenotype and frequent mutation or loss of tumor suppressor genes including Ikzf1 (Ikaros), with targeted deletion Ikzf1 being sufficient to transform Lmo2-dependent leukemias to Lmo2-independence. Moreover, we found that the related transcription factor TAL1 was dispensable in several human T-ALL cell lines that contain SIL-TAL1 chromosomal deletions driving its overexpression, indicating that evolution to oncogene independence can also occur in human T-ALL. Together these results indicate an evolution of oncogene addiction in murine and human T-ALL and show that loss of Ikaros is a mechanism that can promote self-renewal of T-ALL lymphoblasts in the absence of an initiating oncogenic transcription factor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Weinstein IB. Cancer. addiction to oncogenes–the Achilles heal of cancer. Science. 2002;297:63–64.
Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2:251–65.
Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207.
Jonkers J, Berns A. Oncogene addiction: sometimes a temporary slavery. Cancer Cell. 2004;6:535–8.
Karlsson A, Giuriato S, Tang F, Fung-Weier J, Levan G, Felsher DW. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood. 2003;101:2797–803.
Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35:975–83.
Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–8.
Van Vlierberghe P, van Grotel M, Beverloo HB, Lee C, Helgason T, Buijs-Gladdines J, et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood. 2006;108:3520–9.
Wu L, Xu Y, Wang Q, Ruan C, Drexler HG, Wu D, et al. High frequency of cryptic chromosomal rearrangements involving the LMO2 gene in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100:e233–6.
Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87.
Jevremovic D, Roden AC, Ketterling RP, Kurtin PJ, McPhail ED. LMO2 is a specific marker of T-Lymphoblastic Leukemia/Lymphoma. Am J Clin Pathol. 2016;145:180–90.
Matthews JM, Lester K, Joseph S, Curtis DJ. LIM-domain-only proteins in cancer. Nat Rev Cancer. 2013;13:111–22.
Chambers J, Rabbitts TH. LMO2 at 25 years: a paradigm of chromosomal translocation proteins. Open Biol. 2015;5:150062.
McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–83.
Curtis DJ, McCormack MP. The molecular basis of Lmo2-induced T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2010;16:5618–23.
McCormack MP, Shields BJ, Jackson JT, Nasa C, Shi W, Slater NJ, et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood. 2013;122:2093–103.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63.
Garcia-Ramirez I, Bhatia S, Rodriguez-Hernandez G, Gonzalez-Herrero I, Walter C, Gonzalez de Tena-Davila S, et al. Lmo2 expression defines tumor cell identity during T-cell leukemogenesis. EMBO J. 2018;37:e98783.
Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–8.
Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–58.
Kim WI, Wiesner SM, Largaespada DA. Vav promoter-tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp Hematol. 2007;35:1231–9.
Takiguchi M, Dow LE, Prier JE, Carmichael CL, Kile BT, Turner SJ, et al. Variability of inducible expression across the hematopoietic system of tetracycline transactivator transgenic mice. PLoS ONE. 2013;8:e54009.
Bolotin DA, Poslavsky S, Davydov AN, Frenkel FE, Fanchi L, Zolotareva OI, et al. Antigen receptor repertoire profiling from RNA-seq data. Nat Biotechnol. 2017;35:908–11.
Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–64.
Kathrein KL, Chari S, Winandy S. Ikaros directly represses the notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem. 2008;283:10476–84.
Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR, et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006;26:209–20.
Sontani Y, Chapman G, Papathanasiou P, Dunwoodie S, Goodnow CC, Hoyne GF. Cooperation between somatic Ikaros and Notch1 mutations at the inception of T-ALL. Leuk Res. 2011;35:1512–9.
Witkowski MT, Cimmino L, Hu Y, Trimarchi T, Tagoh H, McKenzie MD, et al. Activated Notch counteracts Ikaros tumor suppression in mouse and human T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:1301–11.
Geimer Le Lay AS, Oravecz A, Mastio J, Jung C, Marchal P, Ebel C, et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal. 2014;7:ra28.
Chen S, Nagel S, Schneider B, Kaufmann M, Meyer C, Zaborski M, et al. Novel non-TCR chromosome translocations t(3;11)(q25;p13) and t(X;11)(q25;p13) activating LMO2 by juxtaposition with MBNL1 and STAG2. Leukemia. 2011;25:1632–5.
Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–9.
McCormack MP, Curtis DJ. The thymus under siege: Lmo2 induces precancerous stem cells in a mouse model of T-ALL. Cell Cycle. 2010;9:2267–8.
Palomero T, Odom DT, O’Neil J, Ferrando AA, Margolin A, Neuberg DS, et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 2006;108:986–92.
Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–21.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4.
Sun L, Crotty ML, Sensel M, Sather H, Navara C, Nachman J, et al. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin Cancer Res. 1999;5:2112–20.
Rebollo A, Schmitt C. Ikaros, Aiolos and Helios: transcription regulators and lymphoid malignancies. Immunol Cell Biol. 2003;81:171–5.
Yuan T, Yang Y, Chen J, Li W, Li W, Zhang Q, et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: a novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia. 2017;31:2355–64.
Takanashi M, Yagi T, Imamura T, Tabata Y, Morimoto A, Hibi S, et al. Expression of the Ikaros gene family in childhood acute lymphoblastic leukaemia. Br J Haematol. 2002;117:525–30.
Marcais A, Jeannet R, Hernandez L, Soulier J, Sigaux F, Chan S, et al. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res. 2010;34:426–9.
Kastner P, Chan S. Role of Ikaros in T-cell acute lymphoblastic leukemia. World J Biol Chem. 2011;2:108–14.
Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat Genet. 2011;43:673–8.
Mitchell JL, Yankee TM. Variations in mRNA and protein levels of Ikaros family members in pediatric T cell acute lymphoblastic leukemia. Ann Transl Med. 2016;4:363.
Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–64.
Bellavia D, Mecarozzi M, Campese AF, Grazioli P, Talora C, Frati L, et al. Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 2007;26:1670–80.
Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–41.
Dudgeon C, Chan C, Kang W, Sun Y, Emerson R, Robins H, et al. The evolution of thymic lymphomas in p53 knockout mice. Genes Dev. 2014;28:2613–20.
Lopez-Nieva P, Santos J, Fernandez-Piqueras J. Defective expression of Notch1 and Notch2 in connection to alterations of c-Myc and Ikaros in gamma-radiation-induced mouse thymic lymphomas. Carcinogenesis. 2004;25:1299–304.
Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–99.
Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14:1073–83.
Marke R, van Leeuwen FN, Scheijen B. The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2018;103:565–74.
Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–49.
Deane JE, Mackay JP, Kwan AH, Sum EY, Visvader JE, Matthews JM. Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4. EMBO J. 2003;22:2224–33.
El Omari K, Hoosdally SJ, Tuladhar K, Karia D, Vyas P, Patient R, et al. Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex. Blood. 2011;117:2146–56.
Appert A, Nam CH, Lobato N, Priego E, Miguel RN, Blundell T, et al. Targeting LMO2 with a peptide aptamer establishes a necessary function in overt T-cell neoplasia. Cancer Res. 2009;69:4784–90.
Nam CH, Lobato MN, Appert A, Drynan LF, Tanaka T, Rabbitts TH. An antibody inhibitor of the LMO2-protein complex blocks its normal and tumorigenic functions. Oncogene. 2008;27:4962–8.
Tanaka T, Sewell H, Waters S, Phillips SE, Rabbitts TH. Single domain intracellular antibodies from diverse libraries: emphasizing dual functions of LMO2 protein interactions using a single VH domain. J Biol Chem. 2011;286:3707–16.
Milton-Harris L, Jeeves M, Walker SA, Ward SE, Mancini EJ. Small molecule inhibits T-cell acute lymphoblastic leukaemia oncogenic interaction through conformational modulation of LMO2. Oncotarget. 2020;11:1737–48.
Acknowledgements
The authors thank Alfred Medical Research and Education Precinct (AMREP) animal services and Walter and Eliza Hall Institute (WEHI) Bioservices and for mouse husbandry, Fiona Waters and the WEHI Central Microinjection Service for transgenic mouse generation and the AMREP and WEHI Flow Cytometry Facilities. We thank Cedric Tremblay for antibodies and discussions and Helen Mitchell and Wei Shi for assistance with RNA sequencing and bioinformatics. We thank Charles de Bock, Chris Riffkin, David Huang and Sandra Mifsud for cell lines. This work was supported by project grants (1003391 and 1104145 to MPM), a Program grant (1113577 to WSA), a Fellowship (1058344 to WSA), and the Independent Research Institute’s Infrastructure Support Scheme from the Australian Government’s National Health and Medical Research Council (NHMRC), the Fund for Scientific Research Flanders (LD and PVV), the Ghent University Research Fund and the European Research Council (StG-639784 to PVV), a grant-in-aid from the Cancer Council of Victoria, a Future Fellowship from the Australian Research Council (MPM) and a Victorian State Government Operational Infrastructure Support grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Abdulla, H., Vo, A., Shields, B.J. et al. T-ALL can evolve to oncogene independence. Leukemia 35, 2205–2219 (2021). https://doi.org/10.1038/s41375-021-01120-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-021-01120-9