Abstract
Hashimoto’s disease (HD) is one of the major clinical subtypes of autoimmune thyroid disease. Both environmental and genetic factors contribute to the pathogenesis of HD. Previous evidence has shown that both IRAK2 and TLR10 are potential candidate susceptibility genes for HD. In this study, a total of 3654 Chinese women, including 973 HD cases and 2681 healthy controls, were recruited. Thirty-three tag single nucleotide polymorphisms (SNPs) in IRAK2 and TLR10 were genotyped. Genetic association analyses at both the single-marker and haplotype levels were performed. Gene-by-gene interaction analyses were also conducted in case-only samples, as well as eQTL analyses for significant SNPs based on data extracted from the GTEx database. We identified that two SNPs, rs165501 (OR = 1.20, P = 0.0008, IRAK2) and rs10004195 (OR = 1.23, P = 0.0001, TLR10), were identified to be significantly associated with HD. Rs10004195 was significantly associated with the gene expression of TLR10 in human pituitary tissues (P = 2.00 × 10−4), while rs165501 was significantly associated with the expression of IRAK2 in human thyroid tissues (P = 3.10 × 10−6). No significant results were obtained in the gene-by-gene interaction analyses. Our findings suggest that both IRAK2 and TLR10 play important roles in the onset and development of HD.
Similar content being viewed by others
Introduction
Autoimmune thyroid disease (AITD) constitutes 30% of all autoaggressive disorders, including two main clinical subtypes: Graves’ disease (GD) and Hashimoto’s disease (HD) [1]. HD is more prevalent in women, affecting approximately 5% of the population at some point in their lives [2]. No symptoms are present in the early stage of HD, but painless goiter, hypothyroidism (weight gain, fatigue, constipation, depression, and general pain) and thyroid atrophy develop with the progress of the disease. Morphologically, HD consists of a gradual atrophy of the thyroid tissue following gland invasion with lymphocytic cells, follicular atrophy, and hyperemia accompanied by oncocytic metaplasia of the follicular cells [3]. Thyroid lymphoma is one of the potential complications of HD, but the pathogenesis linking these diseases remains unclear [4]. While both environmental and genetic factors contribute to the pathogenesis of HD, studies of monozygotic twins and CTLA-4 gene polymorphisms have presented strong evidence for a largely genetic etiology of HD [5, 6]. Although these studies confirmed genetic components of HD [7], the etiology and pathogenesis, including finding susceptibility genes for HD, still urgently requires investigation.
With the development of genetic analysis and high-throughput sequencing, more and more susceptibility variants of complex diseases have been identified [8,9,10,11,12,13,14]. Scientists have provided supportive evidence for the polygenic nature of HD and identified some genes, such as SH2B3, associated with HD etiology [15]. Recently, a study investigating polymorphisms in 85 Korean AITD patients and 279 healthy control subjects suggested that Toll-like receptor 10 (TLR10) polymorphisms may contribute to the pathogenesis of AITD [16]. Moreover, TLR10 mRNA was significantly increased in the peripheral blood mononuclear cells of AITD patients compared with those of controls [17], further strengthening the association between TLR10 and AITD. However, these results can explain only a small portion of HD pathogenesis, and the relationship between TLR10 and HD remains unknown. TLRs are a group of transmembrane proteins in humans [18] that act as the primary receptors of innate immunity and are major factors in the pathogenesis of inflammatory diseases, injuries, and cancers. TLR10 is encoded by the TLR10 gene, which is located on chromosome 4p14; it is distinct from other TLRs, having an anti-inflammatory function, and is associated with prostate cancer, asthma and IgA nephropathy [19,20,21]. However, reports of possible associations between TLR10 polymorphisms and HD have been rare to date. TLRs trigger an innate immune response by activating signaling pathways dependent on interleukin-1 receptor-associated kinase (IRAK), and IRAK2 is critical in late-phase TLR responses and the initial responses to TLR stimulation [22, 23]. The IRAK2 protein, which is encoded by the IRAK2 gene located at 3p25.3, is one of two putative serine/threonine kinases that become associated with the interleukin-1 receptor (IL1R) upon stimulation. A previous study has shown that sequence variants in the IRAK1-MECP2 region confer susceptibility to the AITDs GD and HD [24]. However, to the best of our knowledge, no report exists of suspicious associations between IRAK2 and HD or AITD. The information presented here suggests that polymorphisms in TLR10 and IRAK2 may contribute to the pathogenesis of HD, and this possibility requires further study.
With evidence of significant associations between TLR10 and AITD in Korean children [16], HD, as one type of AITD, might also be associated with the TLR10 gene. Therefore, the contributions of TLR10 and IRAK2, a key regulator of the TLR family, to HD risk deserve exploration and study. To further investigate the associations of TLR10 and IRAK2 with the risk of HD, we performed a hospital-based case-control study to further identify the associations of TLR10 and IRAK2 with the risk of HD in Han Chinese individuals. These genetic etiology and pathogenesis data provide clues for understanding the association between TLR10/IRAK2 and HD, and this research will ensure reductions in HD-related morbidity and mortality.
Materials and methods
Study subjects
In the present study, a total of 973 women with HD and 2681 healthy women, controls without any systematic disease, were recruited from the Second Affiliated Hospital of Xi’an Jiaotong University and the Northwest Women and Children Hospital between June 2012 and August 2017. All patients were diagnosed with HD based on an enlarged thyroid, characteristic ultrasound signs (hypoechogenicity and nonhomogeneous texture) and a high level of either antithyroid peroxidase or antithyroglobulin, with or without clinical and biochemical hypothyroidism. Those with a reported history of thyroid cancer and/or previous thyroid surgery were excluded from the study. The healthy controls had normal thyroid functions and no ultrasound changes in the thyroid, and they were negative for thyroid autoantibodies. Exclusion criteria included the existence of any comorbid cardiac, autoimmune, infectious, musculoskeletal or malignant disease, or a recent history of operation or trauma. All participants were unrelated Han Chinese individuals, and the case and control groups were matched by age. The Roche Diagnostics Cobas 6000 E601 Module Immunochemistry Analyzer (Roche, Basal, Switzerland) was used for thyroid function tests, and self-administered questionnaires were used to collect demographic data. The characteristics of our study subjects are shown in Table 1. No significant differences were observed in age, smoking status, or drinking status between the HD cases and healthy controls. Significant differences were found in family history and several clinical parameters (Table 1). The study protocol was approved by the Medical Ethics Committee of Xi’an Jiaotong University in accordance with the ethical guidelines of the Declaration of Helsinki of 1975 (revised in 2008). Written informed consent was obtained from the participants.
SNP selection and genotyping
Candidate SNPs were selected based on 1000 Genomes Chinese Han Beijing population (CHB) data. We searched for all SNPs with a minor allele frequency (MAF) ≥ 0.05 within the regions of the IRAK2 and TLR10 genes. Then, MAF ≥ 0.05 together with r2 ≥ 0.5 for IRAK2 and r2 ≥ 0.7 for TLR10 were used as cutoff criteria during tag SNP selection, which generated 17 and 16 tag SNPs within the IRAK2 and TLR10 genes, respectively. General information about these 33 selected SNPs is summarized in Supplemental Table S1. Genomic DNA was extracted from peripheral blood leukocytes according to the manufacturer’s protocol (Genomic DNA Kit, Axygen Scientific Inc., California, USA). Genotyping was performed for all SNPs using the Sequenom MassARRAY RS1000 system (Sequenom, San Diego, California, USA). The results were processed using Typer Analyser software, and genotype data were generated from the samples [25]. Case and control status were blinded during all genotyping processes for quality control [26]. Five percent of the samples were randomly repeated for genotyping, and the results were the same as before.
Statistical analyses
χ2 tests were performed for each marker to evaluate the genetic risk of HD contribution by single markers. Linkage disequilibrium (LD) blocks were constructed and haplotype-based association analyses were performed by Plink [27]. Potential epistatic effects were evaluated by case-only analyses [28]. To minimize the effects of multiple comparisons, we analyzed only those SNP pairs that included at least one significant hit in the single-marker-based analyses. In addition, we conducted expression quantitative trait loci (eQTL) analyses on those SNPs significantly associated with HD. eQTL data from multiple human tissues were extracted from the GTEx database [29]. In general, Bonferroni corrections were applied to correct for multiple comparisons. For the single-marker-based analyses, the P value threshold used was 0.05/33 ≈ 0.0015. In addition, we also examined the distributions of some clinical indicators of thyroid hormonal status and antithyroid antibodies in patients (free T3, free T4, TSH, anti-TPO and anti-TG) with HD in relation to the significant SNPs. Analysis of variance (ANOVA) was performed to test the significance of the differences in these clinical indicators among the different genotypes. To investigate the potential functional consequences of the two significant SNPs, we utilized bioinformatics tool RegulomeDB (http://www.regulomedb.org/) [30]. RegulomeDB is a database annotating SNPs with known and predicted regulatory elements information integrated from GEO, the ENCODE project, and published literature.
Results
All our candidate SNPs were in Hardy-Weinberg equilibrium in the control samples (Supplemental Table S2). Two SNPs, rs165501 (OR = 1.20, P = 0.0008, IRAK2) and rs10004195 (OR = 1.23, P = 0.0001, TLR10), survived after Bonferroni correction in the single-marker-based association analyses (Table 2 and Supplemental Table S3). Seven LD blocks were constructed (3 for IRAK2 and 4 for TLR10, Supplemental Figs. S1 and S2). A 3-SNP haplotype of TLR10, rs79030744-rs10004195-rs568924323, was identified to be significantly associated with HD disease status (χ2 = 66.88, P = 1.99 × 10−14, Table 3 and Supplemental Table S4, P value threshold was 0.007). A total of 32 SNP pairs including either rs165501 or rs10004195 between IRAK2 and TLR10 were tested for potential epistatic effects. Although several SNP pairs were identified to be nominally significant, none of them survived after Bonferroni correction (Supplemental Table S5, P value threshold was 0.002).
We examined the distributions of the clinical indicators of thyroid hormonal status and antithyroid antibodies in HD patients in relation to rs165501 and rs10004195. As shown in Table 4, the most significant result was obtained for free T4 and rs10004195 in HD patients (P = 0.03). However, this level of significance could not survive after multiple comparison correction. Thus, no significant findings were identified for clinical indicators in HD patients in relation to the two significant SNPs.
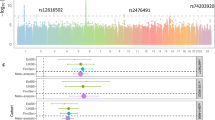
Based on gene expression data extracted from GTEx, both rs165501 and rs10004195 were identified to be eQTLs in specific human tissues (Supplemental Tables S6 and S7, threshold of P values was 5 × 10−4). Rs10004195 was found to be significantly associated with the gene expression of TLR10 in human pituitary tissues (P = 2.00 × 10−4), while rs165501 was significantly associated with the expression of IRAK2 in human thyroid tissues (P = 3.10 × 10−6). Specifically, the C allele of rs165501 was significantly associated with an elevated gene expression level of IRAK2 (Fig. 1). We have also examined the eQTL signals for rs10004195 which achieved genome-wide significance, and the results were summarized in Supplemental Table S8. Rs10004195 was also associated with the expression of some other TLRs including TLR1 and TLR6, in the GTEx database. In the present study, although we have applied SNP-gene mapping strategy based on physical location, SNP rs10004195 was also functional related to some other genes located within ±1 Mb window. RegulomeDB has its own score system and a lower score for a SNP often indicates more functional significance. No results were obtained for rs165501. For rs10004195, we get the score of 1f. This SNP located at two important functional regions: DNasel Hypersensitivity region and H3K27Ac mark region.
Discussion
In this study, we identified significant association signals between our candidate loci, IRAK2/TLR10, and HD disease status. The SNP rs10004195 in TLR10 was identified to be significantly associated with HD, and the odds of A allele in HD patients was averagely 23% higher compared to the controls in our Chinese sample. This SNP was reported to be associated with AITD in a study of Korean children (including 85 AITD patients and 279 healthy controls) conducted by Cho et al. [16]. The A allele of rs10004195 was also reported as a risk allele but with a much larger OR (2.8) in this previous study than in ours. This difference might be partly due to the difference in the sample sizes of the two studies. With a much larger sample size, our estimation for the OR of the risk allele was more accurate with less uncertainty. In addition, to the best of our knowledge, an association signal of rs165501 in IRAK2 with HD has never been reported before. A previous study identified a significant association between IRAK1 and AITD [24], but the association of IRAK2 and HD has never previously been reported in any population-based study. In our study, the odds of C allele of rs165501 was averagely 20% higher in HD patients than in controls. Replication studies based on other populations are needed in the future to validate our findings of the association between IRAK2 and HD or AITD.
Combined with eQTL data extracted from the GTEx database, we explored the potential effects of SNPs that were significantly associated with HD disease status on gene expression levels. Both significant hits showed significant eQTL signals in specific human tissues, and the eQTL pattern of rs165501 was particularly interesting. Our data showed that this SNP could significantly affect the gene expression of IRAK2 in human thyroid tissue, which is a tissue directly related to HD and AITD. The C allele of this SNP seemed to be related to elevated IRAK2 expression. Notably, this C allele is also the risk allele for HD status. Early studies have shown that downregulated expression of IRAK2 could inactivate the NF-κB system [31]. A previous study has reported that HD with chronic inflammation causes a distinct increase in the levels of cytokines and other inflammatory mediators, such as IL-2, INF-gamma, IL-12 and IL-18 [32]. Activation of NF-κB induces the expression of immune response genes [33], including IL-12, which was induced in HD. Coincidentally, the involvement of NF-κB and IL-6 in HD can be seen in a recent study, which indicated that NFKB1 was associated with HD through modulating IL-6 serum levels [34]. Another study demonstrated that the expression of NF-κB and IL-6 was upregulated in the thyroid tissues of adults with HD [35]. Thus, the participation of NF-κB in HD can be confirmed. Interestingly, a genetic variant of IRAK2 can increases NF-κB activity through promoting TRAF6 ubiquitination, and the downstream regulators of NF-κB and IL-6 were excessively activated when IRAK2 was overexpressed [36]. In HD patients, a significantly increased prevalence of variants in the IRAK1-MECP2 region was found [24]. Moreover, our findings on the genetic risk of rs165501 for HD and its eQTL features indicated that this SNP might affect the onset of HD, and this effect was mediated by its effect on the gene expression of IRAK2. Considering the abovementioned information, it is reasonable to hypothesize that IRAK2 plays an essential role in HD through regulating NF-κB and its downstream targets, IL-6 and IL-12. Therefore, a better understanding of the role of IRAK2 in the development of HD may allow early identification of individuals at risk and may even establish novel therapeutic targets in the future.
The evidence has shown a potential biological and functional link between the proteins encoded by IRAK2 and TLR10 [23, 24]. Therefore, we hypothesized that this biological link might be reflected at the level of DNA variations. We conducted a case-only analysis to model the potential gene-by-gene interaction between IRAK2 and TLR10. SNPs from these two loci were expected to be significantly correlated with each other in the case-only samples when epistasis was present. However, no significant signals were obtained from our analysis. This negative result might be partly due to a lack of statistical power, since a larger sample size is often required for gene-by-gene interaction analysis than for single-marker-based association analysis. More research is still needed in the future to detect potential gene-by-gene interactions between IRAK2 and TLR10.
Our study suffered from several limitations. First, in this study, we included only women as our study subjects. Although this sample selection strategy can be partly justified by the higher prevalence of HD in women, it still produced a potential obstacle to generalization of the study results. Another potential limitation of this study is potential false positive signals due to population stratification. Although it is difficult to draw convincing conclusions only from SNP-based association analysis [37,38,39,40,41,42,43,44], haplotype-based association analyses have validated our findings based on single-marker association analyses. However, as one of the major confounders of genetic epidemiology studies, population stratification might severely affect the study results and cause false positive findings. Thus, the results might need to be applied with caution, especially because the Chinese population contains a large amount of genetic heterogeneity. Nevertheless, in this candidate-gene-based study, we could not perform some of the statistical methods that can be applied to genome-wide association studies, such as genomic control or principle component analyses. However, in our participant recruitment process, we tried to restrict the genetic background of our study subjects by applying some selection criteria, such as filtering out those subjects with a familial migration history within three generations. We believe that this strategy at least partly addressed some of the confounding effects of population stratification.
In conclusion, we identified significant association signals between HD disease status and two relevant loci, IRAK2 and TLR10, in the Han Chinese population. Our findings suggested that both IRAK2 and TLR10 play important roles in the onset and development of HD. Given the complex pattern of HD etiology, its potential underlying genetic heterogeneity and its mechanism of chronic inflammation, wider replications in different ethnic samples and further functional analysis are warranted in the pursuit of comprehensive knowledge of the contributions of IRAK2 and TLR10 to the development of HD.
References
Brown RS. Autoimmune thyroid disease: unlocking a complex puzzle. Curr Opin Pediatr. 2009;21:523–8.
Pyzik A, Grywalska E, Matyjaszek-Matuszek B, Roliński J. Immune disorders in Hashimoto’s thyroiditis: what do we know so far? J Immunol Res. 2015;2015:979167.
Pearce EN, Farwell AP, Braverman LE. Thyroiditis. N Engl J Med. 2003;348:2646–55.
Noureldine SI. Tufano RPAssociation of Hashimoto’s thyroiditis and thyroid cancer. Curr Opin Oncol. 2015;27:21–5.
Chistiakov DA. Immunogenetics of Hashimoto’s thyroiditis. J Autoimmune Dis. 2005;2:1–21.
Kavvoura FK, Akamizu T, Awata T, Ban Y, Chistiakov DA, Frydecka I, et al. Cytotoxic T-lymphocyte associated antigen 4 gene polymorphisms and autoimmune thyroid disease: a meta-analysis. J Clin Endocrinol Metab. 2007;92:3162–70.
Wiersinga WM. Clinical relevance of environmental factors in the pathogenesis of autoimmune thyroid disease. Endocrinol Metab. 2016;31:213–22.
Chen G, Guan F, Lin H, Li L, Fu D. Genetic analysis of common variants in the HDAC2 gene with schizophrenia susceptibility in Han Chinese. J Hum Genet. 2015;60:479–84.
Guan F, Li L, Qiao C, Chen G, Yan T, Li T, et al. Evaluation of genetic susceptibility of common variants in CACNA1D with schizophrenia in Han Chinese. Sci Rep. 2015;5:12935.
Guan F, Niu Y, Zhang T, Liu S, Ma L, Qi T, et al. Two-stage association study to identify the genetic susceptibility of a novel common variant of rs2075290 in ZPR1 to type 2 diabetes. Sci Rep. 2015;6:29586.
Guan F, Wei S, Feng J, Zhang C, Xing B, Zhang H, et al. Association study of a new schizophrenia susceptibility locus of 10q24.32-33 in a Han Chinese population. Schizophr Res. 2012;138:63–8.
Guan F, Wei S, Zhang C, Zhang H, Zhang B, Xing B, et al. A population-based association study of 2q32.3 and 8q21.3 loci with schizophrenia in Han Chinese. J Psychiatr Res. 2012;47:712–7.
Guan F, Zhang B, Yan T, Li L, Liu F, Li T, et al. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr Res. 2014;152:97–104.
Guan F, Zhang C, Wei S, Zhang H, Gong X, Feng J, et al. Association of PDE4B polymorphisms and schizophrenia in Northwestern Han Chinese. Hum Genet. 2012;131:1047–56.
Barić A, Brčić L, Gračan S, Torlak Lovrić V, Gunjača I, Šimunac M, et al. Association of established hypothyroidism-associated genetic variants with Hashimoto’s thyroiditis. J Endocrinol Invest. 2017;40:1061–7.
Cho WK, Jang JP, Choi EJ, Jeon YJ, Jung IA, Kim SH, et al. Association of Toll-Like Receptor 10 polymorphisms with autoimmune thyroid disease in Korean children. Thyroid. 2014;25:250–6.
Peng S, Li C, Wang X, Liu X, Han C, Jin T, et al. Increased toll-like receptors activity and TLR ligands in patients with autoimmune thyroid diseases. Front Immunol. 2016;7:578.
Hess NJ, Jiang S, Li X, Guan Y, Tapping RI. TLR10 is a B cell intrinsic suppressor of adaptive immune responses. J Immunol. 2017;198:699–707.
Sun J, Wiklund F, Hsu FC, Bälter K, Zheng SL, Johansson JE, et al. Interactions of sequence variants in interleukin-1 receptor-associated kinase4 and the Toll-like receptor 6-1-10 gene cluster increase prostate cancer risk. Cancer Epidemiol Biomark Prev. 2006;15:480–5.
Lazarus R, Raby BA, Lange C, Silverman EK, Kwiatkowski DJ, Vercelli D, et al. TOLL-like receptor 10 genetic variation is associated with asthma in two independent samples. Am J Respir Crit Care Med. 2004;170:594–600.
Park HJ, Hahn WH, Suh JS, Kim MJ, Kang SW, Lee JS, et al. Association between toll-like receptor 10 (TLR10) gene polymorphisms and childhood IgA nephropathy. Eur J Pediatr. 2011;170:503–9.
Meylan E, Tschopp J. IRAK2 takes its place in TLR signaling. Nat Immunol. 2008;9:581–2.
Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, et al. Sequential control of toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–91.
Song RH, Qin Q, Yan N, Muhali FS, Meng S, Tao HS. et al. Variants in IRAK1-MECP2 region confer susceptibility to autoimmune thyroid diseases. Mol Cell Endocrinol. 2015;399:244–9.
Guan F, Zhang T, Li L, Fu D, Lin H, Chen G, et al. Two-stage replication of previous genome-wide association studies of AS3MT-CNNM2-NT5C2 gene cluster region in a large schizophrenia case-control sample from Han Chinese population. Schizophr Res. 2016;176:125–30.
Guan F, Lin H, Chen G, Li L, Chen T, Liu X, et al. Evaluation of association of common variants in HTR1A and HTR5A with schizophrenia and executive function. Sci Rep. 2016;6:38048.
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Yang Q, Khoury MJ, Sun F, Flanders WD. Case-only design to measure gene-gene interaction. Epidemiology. 1999;10:167–70.
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7.
Muzio M, Ni J, Feng P. Dixit VMIRAK. IRAK(Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–5.
Phenekos C, Vryonidou A, Gritzapis AD, Baxevanis CN, Goula M, Papamichail M. Th1 and Th2 serum cytokine profiles characterize patients with Hashimoto’s thyroiditis (Th1) and Graves’ disease (Th2). Neuroimmunomodulation. 2004;11:209–13.
Böhrer H, Qiu F, Zimmermann T, Zhang Y, Jllmer T, Männel D, et al. Role of NFkappaB in the mortality of sepsis. J Clin Invest. 1997;100:972–85.
Koc A, Batar B, Celik O, Onaran I, Tasan E, Sultuybek GK. Polymorphism of the NFKB1 affects the serum inflammatory levels of IL-6 in Hashimoto thyroiditis in a Turkish population. Immunobiology. 2014;219:531–6.
Wang J, Zhang W, Liu H, Wang D, Wang W, Li Y, et al. Parvovirus B19 infection associated with Hashimoto’s thyroiditis in adults. J Infect. 2010;60:360–70.
Zhang W, He T, Wang Q, Li X, Wei J, Hou X, et al. Interleukin-1 receptor-associated kinase-2 genetic variant rs708035 increases NF-kB activity through promoting TRAF6 ubiquitination. J Biol Chem. 2014;289:12507–19.
Guan FL, Zhang TX, Liu XS, Han W, Lin HL, Li L, et al. Evaluation of voltage-dependent calcium channel gamma gene families identified several novel potential susceptible genes to schizophrenia. Sci Rep. 2016;6:24914.
Han W, Zhang T, Ni T, Zhu L, Liu D, Chen G, et al. Relationship of common variants in CHRNA5 with early-onset schizophrenia and executive function. Schizophr Res. 2018;S0920-9964(18) 30613-3.
Jia XD, Zhang TX, Li L, Fu DK, Lin HL, Chen G, et al. Two-stage additional evidence support association of common variants in the HDAC3 with the increasing risk of schizophrenia susceptibility. Am J Med Genet Part B-Neuropsychiatr Genet. 2016;171:1105–11.
Liu XS, Hou Y, Yan TL, Guo YY, Han W, Guan FL, et al. Dopamine D3 receptor-regulated NR2B subunits of N-Methyl-D-Aspartate receptors in the nucleus accumbens involves in morphine-induced locomotor activity. Cns Neurosci Ther. 2014;20:823–9.
Yang HJ, Zhang B, Zhu JL, Liu D, Guan FL, He XJ. 4q22.1 Contributes to bone mineral density and osteoporosis susceptibility in postmenopausal women of Chinese Han population. PLoS ONE. 2013;8:e80165.
Zhang B, Guan FL, Chen G, Lin HL, Zhang TX, Feng JL, et al. Common variants in SLC1A2 and schizophrenia: association and cognitive function in patients with schizophrenia and healthy individuals. Schizophr Res. 2015;169:128–34.
Zhang T, Liu P, Zhang Y, Wang W, Lu Y, Xi M, et al. Combining information from multiple bone turnover markers as diagnostic indices for osteoporosis using support vector machines. Biomarkers. 2019;24:120–6.
Zhang T, Zhu L, Ni T, Liu D, Chen G, Yan Z, et al. Voltage-gated calcium channel activity and complex related genes and schizophrenia: a systematic investigation based on Han Chinese population. J Psychiatr Res. 2018;106:99–105.
Acknowledgements
This research was totally supported by the National Natural Science Foundation of China (number 81701717 and number 81871366). We thank all of the donors for their assistance in accessing collections and their advice and comments during the preparation of this paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, M., Han, W., Zhu, L. et al. IRAK2 and TLR10 confer risk of Hashimoto’s disease: a genetic association study based on the Han Chinese population. J Hum Genet 64, 617–623 (2019). https://doi.org/10.1038/s10038-019-0613-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0613-5
This article is cited by
-
A tissue-level phenome-wide network map of colocalized genes and phenotypes in the UK Biobank
Communications Biology (2022)